| Issue |
Apidologie
Volume 41, Number 6, November-December 2010
|
|
|---|---|---|
| Page(s) | 654 - 656 | |
| DOI | https://doi.org/10.1051/apido/2010009 | |
| Published online | 15 March 2010 | |
Scientific note
Scientific note on mass collection and hatching of honey bee embryos*
Note scientifique sur une méthode de récolte par lots et d’éclosion simultanée d’embryons d’abeilles
Eine wissenschaftliche Notiz über Sammelverfahren und das Schlüpfen großer Zahlen an Embryonen der Honigbiene
USDA-ARS Bee Research Lab, BARC-East Bldg. 476
Beltsville, MD, 20705,
USA
Corresponding author: J.D. Evans, jay.evans@ars.usda.gov
Received:
11
September
2009
Revised:
4
January
2010
Accepted:
7
January
2010
Key words: queen-rearing / RNA virus / germline transformation
Honey bees provide excellent opportunities for studying development, behavior, and defenses against natural parasites and pathogens. They are also a critical component of modern agriculture, through their leading role in pollinating the world’s crops (Aizen and Harder, 2009). Research and diagnostic methods for honey bees require efficient sampling methods for all colony members, often involving a large number of individual bees. As one example, surveys of honey bees for their pathogens require substantial sample sizes within colonies, especially when there is variation across individuals in infection rate or pathogen loads. This need for greater sampling must be balanced by the expenses involved with collecting and screening numerous colony members. In the case of whole-colony surveys for pathogens, mass extractions from adult workers require scaled up amounts of expensive reagents and, as a result, most such surveys have been limited to a small subset of colony members (vanEngelsdorp et al., 2009). Further, collections of adult workers are likely to contain bees of varied ages and levels of pathogen exposure. For pathogens that are inherited vertically (e.g., many viruses, Chen et al., 2006a,b; Yue et al., 2007; de Miranda and Fries, 2008) it can be more informative to collect embryos as an indicator of pathogen risk. When transmission efficiency from queens to their embryos is variable, such collections need to be made on many individual embryos in order to ensure correct diagnoses. Mass removal of viable embryos can also be used as a screening tool for genetic traits of interest including genetic transformation events and, in principle, an efficient first step in programs for in vitro rearing of honey bee workers, queens, and drones. Here we describe a simple method for collecting thousands of honey bee embryos and show that these embryos have higher hatching rates than do embryos collected by more laborious methods.
To mass-collect embryos, we first placed clean drawn-out comb into colonies. To speed queen egg-laying we restricted queens to the box with the recently placed frame using a queen excluder, although conventional queen push-in cages also would have accomplished this. Two days later, frames were collected and brought to the laboratory. The top bar of each frame was then struck onto a benchtop covered with a clean sheet of semi-flexible plastic (36cm × 52cm) at approximately a 30° angle, and with a force equal to a strong knock (Fig. 1a). This was done four times for each side of the frame when embryos were present. Several hundred embryos per frame were funneled into a microcentrifuge tube by folding the plastic sheet and tapping the backside. Surveys of the frame after striking showed > 90% of embryos had fallen to the plastic. Embryos fell from the frame far more readily than did uncapped brood of all ages. We also sampled unmanipulated field colonies by collecting from frames where embryos were observed and where stored nectar was lowest (nectar also falls out with shaking and complicates the collection of embryos).
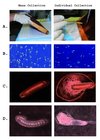 |
Figure 1 Alternate methods for honey bee embryo collection showing (a) collection, (b) embryo state, (c) egg damage, and (d) larval development. |
To test relative viability, an equal number of embryos was collected by knocking the frame and by grafting with a fine needle. Those collected by knocking were funneled into glass Petri dishes (123 embryos split into each of two dishes, 5 minutes total time), while the grafted embryos were placed gently into identical Petri dishes (two dishes with 110 embryos, 30 minutes total time). We checked the viability of embryos by assessing damage and monitoring hatching rates. Embryos were incubated in glass sealed dessicator jars with saturated humidity thanks to a dish containing 15.5% glycerol in water and 0.4% methyl-4-hydroxybenzoate to inhibit microbial growth. Nearly all (115/123) of shaken embryos were intact after 24 hours versus 47/110 embryos carefully removed by a fine needle (Fig. 1b). After 72 hours, 106/123 (86%) of embryos collected by shaking had hatched into healthy larvae versus 34/110 (31%) of the embryos moved with the needle. While mortality of hand-grafted embryos could likely be improved, the strong survival of shaken embryos coupled with the speed of collections makes this a preferable route. A subset of the larvae was reared to pupation without obvious developmental abnormalities.
RNA was extracted en masse from shaken embryos in eight batches of ca. 150 embryos by adding 500 ul TRIzol solution and following standard protocols (Evans et al., 2006). This RNA was quantified at about 30 ng/embryo, giving yields of many ug’s of total RNA from embryos of a single colony. To test whether this RNA might be used as a source for pathogen screening, 2 μg of total RNA was used in two-step RT-PCR reactions with specific viral primers for Deformed wing virus (DWV), an endemic dicistrovirus known to be transmitted vertically by queens (Chen et al., 2006a,b; Yue et al., 2007), Black queen cell virus (BQCV), Kashmir bee virus (KBV), Acute bee paralysis virus (ABPV), and sacbrood virus (SV). With published primers and conditions (Chen et al., 2005), we found strong signals for DWV and BQCV in each of the eight pools of embryos from the sampled colony, and no signal for the other viruses. This suggests that the sampling scheme (150 embryos) is sufficient to generate a consistent view of viral transmission, although more work is needed to determine the benefits of pooling embryos for diagnostics and the scale needed to ensure identification.
Here we show that a snapshot of colony viral loads can be attained by sampling hundreds of embryos quickly and simultaneously. Mass collection of embryos can have uses beyond diagnostics. For one, high amounts of RNA, DNA, proteins or metabolites are often needed for downstream goals such as sequencing or expression assays. In addition, the probabilistic gain of sampling hundreds or even thousands of embryos from each study colony can help in the early search for rare genetic mutants or mating events (e.g., occasional matings with africanized drones), for accurate assessment of the number and relative contributions of mates (e.g., via deep sequencing to identify single-nucleotide polymorphisms linked to specific mates) and for screening queen progeny for rare transformation events or RNA variants found at birth. Finally, mass collection of embryos can facilitate techniques for preserving important genetic stock (Collins, 2002). Surprisingly, embryos collected by the method described here show high survivorship and normal development, allowing for a novel way of raising honey bee progeny for breeding and experimentation.
References
- Aizen M.A. Harder L.D. (2009) The global stock of domesticated honey bees is growing slower than agricultural demand for pollination, Curr. Biol. 19, 915–918. [CrossRef] [PubMed] [Google Scholar]
- Chen Y., Evans J. Feldlaufer M. (2006) Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera, J. Invertebr. Pathol. 92, 152–159. [CrossRef] [PubMed] [Google Scholar]
- Chen Y.P., Pettis J.S., Collins A. Feldlaufer M.F. (2006) Prevalence and transmission of honeybee viruses, Appl. Environ. Microbiol. 72, 606–611. [CrossRef] [PubMed] [Google Scholar]
- Chen Y.P., Pettis J.S., Feldlaufer M.F. (2005) Detection of multiple viruses in queens of the honey bee, Apis mellifera L., J. Invertebr. Pathol. 90, 118–121. [CrossRef] [PubMed] [Google Scholar]
- Collins A.M. (2002) Collection of honey bee eggs for cryopreservation, J. Apic. Res. 41, 89–95. [Google Scholar]
- de Miranda J.R., Fries I. (2008) Venereal and vertical transmission of deformed wing virus in honeybees (Apis mellifera L.), J. Invertebr. Pathol. 98, 184–189. [CrossRef] [PubMed] [Google Scholar]
- Evans J.D., Aronstein K., Chen Y.P., Hetru C., Imler J.-L., Jiang H., Kanost M., Thompson G.J., Zou Z., Hultmark D. (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera, Insect Mol. Biol. 15, 645–656. [CrossRef] [PubMed] [Google Scholar]
- vanEngelsdorp D., Evans J.D., Saegerman C., Mullin C., Haubruge E., Nguyen B.K., Frazier M.A., Frazier J., Cox-Foster D., Chen Y., Underwood R., Tarpy D.R., Pettis J.S. (2009) Colony collapse disorder: A descriptive study, PLoS ONE 4, 8. [Google Scholar]
- Yue C., Schroder M., Gisder S., Genersch E. (2007) Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera), J. Gen. Virol. 88, 2329–2336. [CrossRef] [PubMed] [Google Scholar]
© INRA/DIB-AGIB/EDP Sciences, 2010
All Figures
 |
Figure 1 Alternate methods for honey bee embryo collection showing (a) collection, (b) embryo state, (c) egg damage, and (d) larval development. |
| In the text | |


