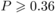| Issue |
Apidologie
Volume 41, Number 4, July-August 2010
|
|
|---|---|---|
| Page(s) | 476 - 487 | |
| DOI | https://doi.org/10.1051/apido/2009083 | |
| Published online | 14 January 2010 | |
Original article
Pollen amino acids and flower specialisation in solitary bees*
Acides aminés du pollen et spécialisation florale chez les abeilles solitaires
Aminosäuren im Pollen und Blütenspezialisierung bei solitären Bienen
Department of Animal Ecology and Tropical Biology University of Würzburg,
Germany
Corresponding author: N. Blüthgen, bluethgen@biozentrum.uni-wuerzburg.de
Received:
16
July
2009
Revised:
20
October
2009
Pollen nutrient composition could be important in host-plant selection of oligolectic bees. In this study, pollen samples from 142 plant species were analysed separately for water-soluble and protein-bound amino acids. The composition of amino acids varied strongly among plant species, but taxonomically related species had similar compositions. All plant species contained the entire set of essential amino acids, although some in small quantities. Total concentration of free- and protein-bound amino acids was significantly lower in pollen sources used by oligoleges than in other pollen sources. Pollen sources of oligoleges showed a lower concentration of essential amino acids and deviated more strongly from the ideal composition of essential amino acids as determined for honey bees than plants not hosting oligoleges. However, this trend was not confirmed on a cruder phylogenetic plant family level, where pollen chosen by oligolectic bees was similar to other pollen.
Zusammenfassung
Die meisten Bienen ernähren sich ausschließlich von Pollen und Nektar, wobei Pollen die primäre Proteinquelle ihrer Larven darstellt. Während oligolektische Bienen auf den Pollen einer oder mehrerer nah verwandter Pflanzenarten spezialisiert sind, ist das Blütenspektrum polylektischer Bienen breiter. Der Vorteil der Oligolektie ist bisher weitgehend unbekannt, wobei eine Vielzahl von Hypothesen diskutiert wird. Dazu gehören eine höhere Effizienz der Pollenspezialisten beim Sammeln und bei der Verdauung des Pollens, sowie eine Spezialisierung auf Pollen mit höherem Stickstoffgehalt. Unser Ziel war, herauszufinden, ob die Pollenqualität, insbesondere der Anteil der essentiellen Aminosäuren (Abb. 1), für die Wahl bestimmter Pflanzenarten durch oligolektische Bienen verantwortlich sein könnte. Die Aminosäurezusammensetzung der Pollen von 142 Pflanzenarten (Tab. I) zeigte signifikante Unterschiede zwischen Pflanzenfamilien (Abb. 2). Von oligolektischen Bienen genutzter Pollen unterschied sich jedoch in der Komposition nicht signifikant von anderen Pollenarten. Allerdings enthielt der von oligolektischen Bienen genutzte Pollen eine signifikant geringere Konzentration an Aminosäuren (Abb. 3). Zudem zeigte sich eine verminderte Nahrungsqualität bei Pollenquellen oligolektischer Bienen: Die Komposition essentieller Aminosäuren zeigte eine signifikant größere Diskrepanz zu der für Honigbienen als ideal beschriebenen Komposition als die übrigen Pollenarten. Daher könnte spekuliert werden, dass oligolektische Bienen nährstoffärmeren Pollen nutzen, um interspezifische Konkurrenz mit anderen Pollenkonsumenten zu verringern. Hinweise auf tatsächlich verminderte Konkurrenz gibt es jedoch bislang nicht. Der Befund, dass oligolektische Bienen auf qualitativ minderwertigen Pollen spezialisiert sind, ist zudem stark geprägt durch die in der Analyse überrepräsentierten Asteraceen und Lamiaceen. Diese weisen ähnlich geringe Aminosäurekonzentrationen auf. Auf Familienniveau zeigte der von oligolektischen Bienen genutzte Pollen keine signifikant geringere Qualität.
Key words: solitary bees / pollen / amino acids / oligolecty
Mots clés : acides aminés / pollen / abeilles solitaires / oligolectie
Schlüsselwörter: Solitäre Bienen / Pollen / Aminosäuren / Oligolektie
© INRA/DIB-AGIB/EDP Sciences, 2010
1. INTRODUCTION
Most bees feed exclusively on plant pollen and nectar, representing their primary source of protein and other nutrients especially during the larval stage (Westrich, 1990). While oligolectic bees depend on pollen from a single plant species, genus, or family, polylectic bees use a broad spectrum of flowering plants (Cane and Sipes, 2006). Traditionally, it has been assumed that polylecty was the ancestral state in bees (Michener, 1954). Indeed, this proved to be true for the Hemihalictus series in the genus Lasioglossum (Danforth et al., 2003), but recently growing evidence suggests that in the majority of bee lineages generalist species have evolved from oligolectic ancestors (Larkin et al., 2008; Michez et al., 2008).
The advantages of oligolecty remain largely unknown, though several hypotheses have been discussed, above all a higher proficiency of specialised bees when visiting their specific host flowers through evolutionary adaptation (Strickler, 1979; Müller, 2006). On the other hand, host-plant specialisation among bees could have been favoured if it reduced interspecific competition (Thorp, 1969). As all plant species visited by oligoleges are visited by polyleges as well, at least a complete escape from competition seems to be unlikely (Minckley and Roulston, 2006). However, some quantitative extent of competition-avoidance could be achieved by specialising on pollen containing toxic compounds or being less nutritious and therefore visited less frequently or by fewer species. On the other hand, it has been suggested that oligolectic bees specialise on plant pollen with higher nitrogen content, but this hypothesis is lacking in phylogenetically sound evidence so far (Budde et al., 2004). Pollen nutritional value has been judged mostly by its crude protein content (Day et al., 1990), estimated based on pollen nitrogen concentration multiplied by 6.25 (e.g. Rabie et al., 1983). This conversion factor may not be appropriate for pollen (Roulston and Cane, 2000). Moreover, protein content may not adequately reflect the availability and composition of amino acids; two diets containing the same protein content may differ in nutritional value due to a lack or imbalance of essential amino acids (Standifer, 1967).
Insects and other animal taxa have relatively similar basic nutritional requirements, including the spectrum of essential amino acids (De Groot, 1952). It has been demonstrated that dietary protein content is crucial for reproduction, growth and longevity of bees and other insects (Gilbert, 1972; Roulston and Cane, 2002). Preferences for diets with higher amino acid content have been documented in studies on butterflies (Erhardt and Rusterholz, 1998), ants (Blüthgen and Fiedler, 2004), parasitoid wasps (Wäckers, 1999), and honey bees (Alm et al., 1990). The ideal composition of essential pollen amino acids (arginine 11%, histidine 5%, isoleucine 14%, leucine 16 %, lysine 11%, methionine 5%, phenylalanine 9%, threonine 11%, tryptophan 4%, valine 14%) determined for the honey bee, Apis mellifera, by De Groot (1953) were very similar to those of other animals (Nation, 2002). Thus, it can be assumed that bees do not vary significantly in their nutritional requirements concerning relative amino acid composition.
We focused on qualitative as well as quantitative pollen amino acid composition and balance. Our goal was to find out whether the pollen of plants selected by oligolectic bee species differs in its chemical composition compared to the pollen of plants not hosting oligoleges. We tested whether pollen sources of oligoleges contained either a significantly higher or lower (1) total amino acid content or (2) balanced composition of essential amino acids, and (3) deviation from an ideal composition of essential amino acids proposed by De Groot (1953) than plants not hosting oligoleges.
2. MATERIAL AND METHODS
2.1. Pollen collection and analysis
Analysed plant taxa, their assignment to one of the two tested groups (O = pollen hosts of oligolectic bees, N = not hosting oligolectic bees) and the total concentration of free and protein-bound amino acids as well as percentage of essential amino acids (AA = amino acids).
Pollen from 142 plant species was sampled and analysed for its amino acid composition (Tab. I). The nomenclature followed Wisskirchen and Haeupler (1998). Among these plants only five species may not be regularly visited by bees (namely Caltha palustris L., Circaea lutetiana L., Erophila verna L., Sambucus nigra L., Silene latifolia Poir.). However, excluding the plants from the analysis did not affect the overall results. Ninety-one of the sampled species have been either confirmed to be visited by oligoleges through pollen analysis from bee pollen scopa and/or observations (Westrich, 1990; Müller et al., 1997; Müller, 2006) or belong to plant genera known to host oligolectic bees. We included all species belonging to a plant genus visited by oligoleges into this group, as most oligoleges are assumed to be specialised on the genus or family level (Minckley and Roulston, 2006), and observations may not cover all potential pollen host species. This yielded a total of 91 plant species hosting oligoleges and 51 plant species not hosting oligoleges. Twenty-nine plant species hosting oligoleges belonged to the family of Asteraceae and 11 species to the family of Lamiaceae. Such families with high replication may be assumed to be overrepresented in the results on the species level. We therefore present an additional test where amino acid values have been pooled for each of the 41 plant families to check whether patterns were consistent on this crude phylogenetic level (Noligolectic = 22 and Ngeneralised = 26 families, Tab. I). In families containing plants visited and plants not visited by oligoleges, we pooled plants for each category separately, which resulted in seven plant families occurring twice.
So far, most studies analysed bee-collected rather than hand-collected pollen and were based on a few plant taxa only (references in: Roulston and Cane, 2000, but see Wille et al., 1985). The analysis of bee-collected pollen is problematic as bees add substantial amounts of nectar to pollen loads (Müller et al., 2006; Leonhardt et al., 2007). This creates an unknown bias caused by nectar derived sugars accounting for up to 40% of the pollen pellet’s dry weight (Roulston and Cane, 2000). Any analysis of pollen pellets that disregards the added weight of nectar sugars to the pellets greatly underestimates the concentration of proteins in the pollen itself. This bias cannot be removed by a standardized multiplier (Roulston and Buchmann, 2000).
We therefore attempted to discover differences in the pollen nutritive value using hand-collected pollen samples only. For each sample, depending on pollen amount per plant species, pollen from 2–400 flower heads was pooled to yield sufficient amounts for analysis (0.08–9.6 mg). Large samples were subsampled for multiple determinations. As manual grinding of pollen using a mortar and a pestle prior to extraction did not change results in terms of total amino acid content (Wilcoxon; Z = 1.54, P = 0.12, N = 8 plant species) samples were not ground. The overall trend even showed higher contents in untreated pollen (on average 6.15 μg/mg). Each sample was checked for contaminations under a stereo microscope and then frozen at –20 °C until it was prepared for analysis by drying over night at 30 °C. Longer drying did not further decrease pollen dry weight.
Free and protein-bound amino acids were measured separately with an ion exchange chromatograph (Biotronik, amino acid analyser LC 3000). For analysis of water soluble amino acids, usually 3–5 mg (dry weight) pollen was extracted with 100 μL water for 30 min in an ultrasonic bath (EMAG, Emmi 20HC) and afterwards for 60 min in the refrigerator. After centrifugation (15000 g) and membrane filtration for 10 min, the sediment was saved for later analysis of the amino acids in the protein fraction. The supernatant was poured into a new microcentrifuge tube, boiled for 2 min at 100 °C, and cooled in ice to room temperature before a second centrifugation for 5 min. Afterwards, 50 μL of the supernatant was extracted with 10 μL 12.5% 5-sulfosalicylic acid in the refrigerator for 30 min for precipitation of proteins. Ten minutes of centrifugation followed, and 50 μL of the supernatant plus 50 μL thinning buffer were poured into a fresh tube, mixed, and pipetted in a membrane filter (Vecta Spin) before a last centrifugation for 5 min and adjacent measurement in the amino acid analyser.
For analysis of the amino acids of the protein fraction, 200 μL of 6 N HCl3 was added to the sediment, the sample was mixed, boiled for four hours at 100 °C, and cooled to room temperature. 10 min of centrifugation followed. The supernatant was poured into a new tube and evaporated at 100 °C. Afterwards, the sample was re-dissolved in 200 μL of water, immediately cooled to room temperature, and centrifuged again (10 min). Subsequently, 100 μL of the supernatant was mixed with 20 μL 12.5% sulphosalicylic acid and extracted 30 minutes in the refrigerator before short mixing and centrifugation for another 10 minutes. 100 μL of the supernatant and 100 μL sample rarefaction buffer was transferred into a new microcentrifuge tube. All was pipetted through a membrane filter, centrifuged for 5 minutes, and transferred into a new microcentrifuge tube for further rarefaction with sample rarefaction buffer (1:5) before measurement.
The experimental variability of our technique yielded a median coefficient of variation (CV = standard deviation/mean) of 0.383, with a median standard deviation (SD) of 8.52 μg/mg pollen (n = 91 repeatedly measured pollen samples). It is much smaller than the variability between samples of the same species varying in date or place of collection (median CV = 1.084, median SD = 12.16 μg/mg; n = 31 pollen samples of the same species). If pollen from a plant species was analysed in more than one sample, for consistency we included only the sample with the highest pollen dry weight into statistical analyses. However, there was no trend towards higher amino acid concentration in samples higher in weight (paired t-Test, t = 0.66, P = 0.707, n = 91 pairs). We compared total amino acid content yielded with our method to protein content of the same samples analysed in Bradford assays. Our results are linearly correlated (y = 0.76x + 44.61, R2 = 0.91) and slightly higher for each of the plant species analysed. Our results are comparable to those of Standifer (1967).
2.2. Statistical analysis
The composition of pollen amino acids was examined using non-metric multidimensional
scaling (NMDS), employing a Bray-Curtis similarity matrix, two dimensions, and 1000 runs.
Statistics were conducted in R 2.6 (R Development Core Team 2006) using the « metaMDS »
command and 1000 iterations (R-package vegan 1.8.2). Amino acid composition data were
entered as molar proportions (amino acidi [μMol
g−1] / total amino acid concentration [μMol g−1])
based on dry weight. To analyse differences among plant families and between the groups of
plants hosting vs. not hosting oligoleges, analysis of variance using distance matrices («
adonis » command, R-package vegan) was conducted. The balance of the proportions of amino
acids was measured as standardized evenness derived from Simpson’s diversity index:
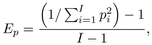 where pi is
the molar proportion of each amino acid i of the total concentration of
I amino acids. EP
approaches 0 for the most imbalanced composition and 1 for a perfectly homogenous
composition with each amino acid occurring in the same proportion. The deviation of
essential pollen amino acid composition from the ideal composition determined for the
honey bee by De Groot (1953) was measured as
Bray-Curtis distances. Mann-Whitney U-tests (two-tailed) were conducted to examine whether
plants hosting oligolectic bees differed in any parameter from plants not known to host
oligoleges. All analyses were performed for total amino acids (free plus protein-bound)
and separately for the free amino acids alone. Moreover, separate analyses were performed
for the whole spectrum of amino acids (see Fig. 1)
and only for the essential ten, namely arginine, histidine, isoleucine, leucine, lysine,
methionine, phenylalanine, threonine, tryptophan, and valine (De Groot, 1952).
where pi is
the molar proportion of each amino acid i of the total concentration of
I amino acids. EP
approaches 0 for the most imbalanced composition and 1 for a perfectly homogenous
composition with each amino acid occurring in the same proportion. The deviation of
essential pollen amino acid composition from the ideal composition determined for the
honey bee by De Groot (1953) was measured as
Bray-Curtis distances. Mann-Whitney U-tests (two-tailed) were conducted to examine whether
plants hosting oligolectic bees differed in any parameter from plants not known to host
oligoleges. All analyses were performed for total amino acids (free plus protein-bound)
and separately for the free amino acids alone. Moreover, separate analyses were performed
for the whole spectrum of amino acids (see Fig. 1)
and only for the essential ten, namely arginine, histidine, isoleucine, leucine, lysine,
methionine, phenylalanine, threonine, tryptophan, and valine (De Groot, 1952).
 |
Figure 1 Amino acid profile of two exemplary plant species: Campanula trachelium and Agrimonia eupatoria. C. trachelium hosts oligolectic bees unlike A. eupatoria. All measured amino acids and their derivates are displayed, separated into free and protein-bound fractions. (Arg = arginine, His = histidine, Ile = isoleucine, Leu = leucine, Lys = lysine, Met = methionine, Phe = phenylalanine, Thr = threonine, Trp = tryptophan, Val = valine, α-AAA = α-aminoadipic acid, α-ABA = α-aminobutyric acid, Ala = alanine, Asn = asparagine, Asp = aspartic acid, β-AIBA = β-aminoisobutyric acid, β-Ala = β-Alanine, Car = carnosine, Citr = citrulline, Cys = cysteine, Cyst = cystathionine, GABA = γ-aminobutyric acid, Gln = glutamine, Glu = glutamic acid, Gly = Glycin, 1-Meth = 1-methylhistidine, 3-Meth = 3-methylhistidine, OH-Pro = hydroxyproline, Orn = ornithine, P-Eta = phosphoethanolamine, Pro = proline, P-Ser = phosphoserine, Ser = serine, Tau = taurine, Tyr = tyrosine). |
3. RESULTS
Plants differed strongly in their composition of pollen amino acids, especially in the proportions of free and protein-bound amino acids (Fig. 1). Closely related plant species plotted together on the ordination diagram, showing similar pollen chemistry (Fig. 2). Differences across families were significant (ADONIS; R2 = 0.677, P < 0.01). However, plant species supporting oligolectic bees did not differ significantly from other plants in overall amino acid composition of pollen (R2 = 0.002, P = 0.58) and are scattered among them (Fig. 2).
 |
Figure 2 Taxonomic signals in pollen amino acids: closely related species often have a similar chemistry. Each symbol denotes one plant species. Plants hosting oligoleges are displayed with « + » plants not known to host them with « o ». Species that plot together are similar in their relative proportions of amino acids (free and protein-bound pooled). Four examples of plant families are highlighted to indicate their similar pollen composition (NMDS, stress = 9.53, Bray-Curtis similarity). |
 |
Figure 3 Amino acids (AA) compared between plants hosting oligolectic bees vs. plants not
known to host oligoleges. Box whisker plots showing median, quartiles and range.
Plants hosting oligoleges showed a significantly lower pollen quality in terms of
total amino acid concentration (Mann-Whitney U-test; Z = − 2.69, P =
0.0072) and total essential amino acids (Z = − 2.64, P = 0.0082),
whereas the fractions of free and free essential amino acids did not show significant
differences (all Z ≤ 0.92, all |
Average amino acid concentrations differed significantly between pollen from plant species
supporting oligolectic bees and pollen collected from plants not hosting oligoleges (Fig.
3). Plants hosting oligoleges showed a significantly
lower pollen quality, both in terms of total amino acid concentration and the fraction of
all essential amino acids. This differentiation was found in the pooled total, but not in
the fraction of free amino acids (Fig. 3). However,
some plant families, namely Asteraceae and Lamiaceae, are overrepresented in the genus-level
sample and thus shape the results on this specific level. When data are pooled at the family
level, no significant differences between plant families visited and families not visited by
oligoleges remained (Mann-Whitney U tests for groups of compounds as in Fig. 3, all Z ≤ 0.35, 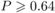 ,
Noligolectic = 22, Ngeneralised = 26).
,
Noligolectic = 22, Ngeneralised = 26).
The balance of amino acids (evenness) did not vary significantly between plants hosting
oligolectic bees and plants not hosting oligoleges. This was true also for each of the
fractions described above (all Z ≤ 0.86, all 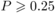 ,
Noligolectic = 91, Ngeneralised = 51 plant species). However, plant
genera hosting oligoleges had a significantly less ideal composition of essential pollen
amino acids on the basis determined by De Groot (1953) for honey bees than the other plants (Z = 2.66, P = 0.008).
The mean (± SD) Bray-Curtis distance between pollen and the ideal composition for pollen
collected by oligolectic bees was 0.179 (± 0.03) and 0.161 ( ± 0.02) for pollen not known
to be collected by oligolectic bees. In particular, plants hosting oligoleges contain a
significantly smaller proportion of valin (Z = 2.58, P = 0.0099), isoleucin
(Z = 3.17, P = 0.002), leucin (Z = 2.08, P = 0.037), and
arginin (Z = 1.98, P = 0.048) but a higher proportion of histidin (Z =
2.65, P = 0.008). However, after phylogenetic correction only the result
for isoleucin remains significant (Z = 2.07, P = 0.039).
,
Noligolectic = 91, Ngeneralised = 51 plant species). However, plant
genera hosting oligoleges had a significantly less ideal composition of essential pollen
amino acids on the basis determined by De Groot (1953) for honey bees than the other plants (Z = 2.66, P = 0.008).
The mean (± SD) Bray-Curtis distance between pollen and the ideal composition for pollen
collected by oligolectic bees was 0.179 (± 0.03) and 0.161 ( ± 0.02) for pollen not known
to be collected by oligolectic bees. In particular, plants hosting oligoleges contain a
significantly smaller proportion of valin (Z = 2.58, P = 0.0099), isoleucin
(Z = 3.17, P = 0.002), leucin (Z = 2.08, P = 0.037), and
arginin (Z = 1.98, P = 0.048) but a higher proportion of histidin (Z =
2.65, P = 0.008). However, after phylogenetic correction only the result
for isoleucin remains significant (Z = 2.07, P = 0.039).
Most sampled species contained the full spectrum of essential amino acids. However, tryptophan had particularly low levels (< 1.0 μMol/g) in more than one-third of the plant species analysed, and methionine was present only in traces in Pastinaca sativa and Erigeron annuus. The total concentration ranged from 0.04 μMol/g dry weight in Silene dioica to 15.8 μMol/g in Corydalis cava.
4. DISCUSSION
Our comparison of pollen amino acid composition showed that closely related species differ only slightly in their proportions of amino acids, suggesting that the profiles are a highly conserved trait. Compositional differences were most obvious between families and orders. Most plant species investigated contained the full spectrum of essential amino acids, albeit some in extremely small quantities. Earlier studies reported that tryptophan was lacking in several pollen species (Auclair and Jamieson, 1948; Roulston and Cane, 2000), partly for plants where tryptophane was detected only in trace amounts in our analysis. However, the strong quantitative limitation of tryptophane and occasionally methionine is evident, and this limitation may be crucial for the development of bees or other pollen feeding insects.
Regarding pollen amino acid concentration at the family level (to compensate overrepresentation of closely related plants), our results are consistent with earlier findings of crude protein or nitrogen contents (Roulston et al., 2000); pollen known to be collected by oligoleges is neither more nor less nutritious than other pollen. On the species level, oligolege pollen hosts contain significantly lower amounts of amino acids. These conflicting findings at the family and species levels may result from the latter being strongly dominated by common plant families, particularly Asteraceae and Lamiacae. Indeed, some plants families are over-proportionally visited by oligoleges, whereas others do not host oligoleges at all. This suggests that evolutionary constraints may have played a major role in host-plant choice of oligoleges (Sedivy et al., 2008).
Host plants of oligoleges showed a poorer match to the ideal composition of essential pollen amino acids determined by De Groot (1953) than other plants. It may thus be possible that oligolectic bees are better adapted to a poorer nutritional quality of their host plants, among many other adaptations to their specific pollen sources. Accordingly, it has been hypothesised that specialist bee species may be more efficient in resource use than related generalists (Strickler, 1979; Dobson and Peng, 1997). Higher efficiency in pollen harvesting can be achieved through behavioural and morphological adaption (Müller and Bansac, 2004). Examples are modification of mouth parts in oligolectic Leioproctus or a specialised hind-leg brush in oligolectic Megachile species (Houston, 1989; Müller and Bansac, 2004). The evolution of such specialised pollen-removal structures evolved several times independently in widely separated taxa, but it is not restricted to oligolectic bees (Thorp, 2000). Nevertheless, Michez et al. (2008) found some evidence that host switches occur more frequently to morphologically similar rather than closely related plants.
Shorter handling time per flower and the ability of oligoleges to remove more pollen per flower than generalists was reported by Strickler (1979) and Cane and Payne (1988). These skills may lead to higher potential reproduction, since more pollen is collected for the brood cells per unit of handling time. However, bees do not adjust pollen provision based on the pollen’s protein content. Roulston and Cane (2002) found the amount of pollen provision to predict larval performance only if, additionally to provision mass, protein content was considered. Besides, some evidence suggests that oligoleges are physiologically better adapted to digestion of their host-plant pollen and can absorb the nutrients present in the pollen of their restricted food source more effectively than other bees (Dobson and Peng, 1997; Praz et al., 2008). This might explain a choice of pollen species with lower total or essential amino acids. However, polylectic bees commonly collect monospecific pollen loads for nest provision (Westrich, 1990) and thus also depend on the suitability of their particular provision. In brood cells containing pollen loads deficient in one or more essential amino acids, larvae would not be able to develop. Thus, it may not be surprising that polyleges select similar or even better pollen qualities.
Adaptation to a certain pollen source may be associated with a cost: a decreased capability to digest other pollen types. Such costs are known to occur in host-specific herbivores (Strauss and Zangerl, 2002) and were recently hypothesised for bees as well (Sedivy et al., 2008). While some studies demonstrated that oligoleges grow well on some non-host pollen (Bohart and Youssef, 1976; Williams, 2003), brood failure has been reported in other investigations (Guirguis and Brindley, 1974; Praz et al., 2008). In some cases toxic compounds may be involved in specialist bees being able to cope better with some pollen species than others (Praz et al., 2008). To our knowledge, no comparative approach of pollen toxins exists so far. If oligolectic bees specialised on pollen that is either deficient in amino acids or contains toxic compounds, this might have led to a competitive advantage in terms of the available pollen quantity and may explain why Asteraceae host large numbers of oligoleges but only few polyleges (Müller and Kuhlmann, 2008). However, effective competitive avoidance has not been demonstrated so far. Most plant species visited by oligoleges are also regularly visited by polylectic bees and other insects, but this does not exclude the possibility of quantitative effects of competitive avoidance. Answering this question would require quantitative surveys of flower visitation and pollen removal rates.
Acknowledgments
We thank Andreas Müller for valuable comments on the manuscript, Dorothea Hahne and Verena Seufert, who patiently assisted with pollen collection and the entire team of the Biodiversity Exploratories for logistic support. This study is part of the Biodiversity Exploratories funded by the Deutsche Forschungsgemeinschaft (LI 150/201, www.biodiversity-exploratories.de).
References
- Alm J., Ohnmeiss T.E., Lanza J., Vriesenga L. (1990) Preference of cabbage white butterflies and honey bees for nectar that contains amino acids, Oecologia 84, 53–57. [CrossRef] [PubMed] [Google Scholar]
- Auclair J.L., Jamieson C.A. (1948) Qualitative analysis of amino acids in pollen collected by bees, Science 108, 357–358. [CrossRef] [PubMed] [Google Scholar]
- Blüthgen N., Fiedler K. (2004) Preferences for sugars and amino acids and their conditionality in a diverse nectar-feeding ant community, J. Anim. Ecol. 73, 155–166. [CrossRef] [Google Scholar]
- Bohart G.E., Youssef N.N. (1976) The biology and behavior of Evylaeus galpinsiae Cockerell (Hymenoptera: Halictidae), Wasmann J. Biol. 34, 185–234. [Google Scholar]
- Budde J., Reckert A., Sporer F., Wink M., Eltz T., Lunau K. (2004) Beiträge zur Evolution der Oligolektie bei solitären Bienen der Gattung Andrena, Entomol. Heute 16, 191–200. [Google Scholar]
- Cane J.H., Payne J.A. (1988) Foraging ecology of the bee Habropoda laboriosa (Hymenoptera: Anthophoridae), an oligolege of blueberries (Ericaceae: Vaccinium) in the southeastern United States, Ann. Entomol. Soc. Am. 81, 419–427. [Google Scholar]
- Cane J.H., Sipes S.D. (2006) Characterizing floral specialization by bees: Analytical methods and a revised lexicon for oligolecty, in: Waser N.M., Ollerton J. (Eds.), Specialization and Generalization in Plant-Pollinator Interactions, University of Chicago Press, Chicago, pp. 99–122. [Google Scholar]
- Danforth B.N., Conway L., Ji S. (2003) Phylogeny of eusocial Lasioglossum reveals multiple losses of eusociallity within a primitively eusocial clade of bees (Hymenoptera: Halictidae), Syst. Biol. 52, 23–36. [CrossRef] [PubMed] [Google Scholar]
- Day S., Beyer R., Mercer A., Ogden S. (1990) The nutrient composition of honeybee-collected pollen in Otago New Zealand, J. Apicult. Res. 29, 138–146. [Google Scholar]
- De Groot A.P. (1952) Amino acid requirements for the growth of the honeybee (Apis mellifera L.), Experentia 8, 1992–1994. [Google Scholar]
- De Groot A.P. (1953) Protein and amino acid requirements of the honey bee (Apis mellifera L.), Phys. Comp. Oec. 3, 197–285. [Google Scholar]
- Dobson H.E.M., Peng Y.-S. (1997) Digestion of pollen components by larvae of the flower-specialist bee Chelostoma florisomne (Hymenoptera: Megachilidae), J. Insect Physiol. 43, 89–100. [CrossRef] [PubMed] [Google Scholar]
- Erhardt A., Rusterholz H.P. (1998) Do peacock butterflies (Inachis io L.) detect and prefer nectar amino acids and other nitrogenous compounds? Oecologia 117, 536–542. [CrossRef] [PubMed] [Google Scholar]
- Gilbert L.E. (1972) Pollen feeding and reproductive biology of Heliconius butterflies, Proc. Natl Acad. Sci. (USA) 69, 1403–1407. [Google Scholar]
- Guirguis G.N., Brindley W.A. (1974) Insecticide susceptibility and response to selected pollens of larval alfalfa leafcutting bees, Megachile pacifica (Panzer) (Hymenoptera: Megachilidae), Environ. Entomol. 3, 691–694. [Google Scholar]
- Houston T.F. (1989) Leioproctus bees associated with Western Australian smoke bushes (Conospermum spp.) and their adaptations for foraging and concealment (Hymenoptera: Colletidae: Paracolletini), Rec. West. Aust. Mus. 14, 275–292. [Google Scholar]
- Larkin L.L., Neff J.L., Simpson B.B. (2008) The evolution of a pollen diet: host choice and diet breath of Andrena bees (Hymenoptera: Andrenidae), Apidologie 39, 133–145. [CrossRef] [EDP Sciences] [Google Scholar]
- Leonhardt S., Dworschak K., Eltz T., Blüthgen N. (2007) Foraging loads of stingless bees and utilisation of stored nectar for pollen harvesting, Apidologie 38, 125–135. [CrossRef] [EDP Sciences] [Google Scholar]
- Michener C.D. (1954) Records and describtions of North American megachilid bees, J. Kansas Entomol. Soc. 27, 65–78. [Google Scholar]
- Michez D., Patiny S., Rasmont P., Timmermann K., Vereecken N.J. (2008) Phylogeny and host-plant evolution in Melittidae s.l. (Hymenoptera: Apoidea), Apidologie 39, 146–162. [CrossRef] [EDP Sciences] [Google Scholar]
- Minckley R.L., Roulston T.H. (2006) Incidental mutualisms and pollen specialization among bees, in: Waser N.M., Ollerton J. (Eds.), Plant-pollinator interactions from specialization to generalization, University of Chicago Press, Chicago, Illinois, USA, pp. 69–98. [Google Scholar]
- Müller A. (2006) Unusual host plant of Hoplitis pici, a bee with hooked bristles on its mouthparts (Hymenoptera: Megachilidae: Osmiini), Eur. J. Entomol. 103, 497–500. [Google Scholar]
- Müller A., Bansac N. (2004) A specialized pollen-harvesting device in western palearctic bees of the genus Megachile (Hymenoptera, Apoidea, Megachilidae), Apidologie 35, 329–337. [CrossRef] [EDP Sciences] [Google Scholar]
- Müller A., Diener S., Schnyder S., Stutz K., Sedivy C., Dorn S. (2006) Quantitative pollen requirements of solitary bees: Implications for bee conservation and the evolution of bee-flower relationships, Biol. Conserv. 130, 604–615. [Google Scholar]
- Müller A., Krebs A., Amiet F. (1997) Bienen – Mitteleuropäische Gattungen, Lebensweise, Beobachtung, Naturbuch Verlag, Augsburg. [Google Scholar]
- Müller A., Kuhlmann M. (2008) Pollen hosts of western palearctic bees of the genus Colletes (Hymenoptera:Colletidae): the Asteraceae paradox, Biol. J. Linn. Soc. 95, 719–733. [CrossRef] [Google Scholar]
- Nation J.L. (2002) Insect physiology and biochemistry, CRC Press, Boca Raton, Florida. [Google Scholar]
- Praz C.J., Müller A., Dorn S. (2008) Specialized bees fail to develop on non-host pollen: do plants chemically protect their pollen? Ecology 89, 795–804. [CrossRef] [PubMed] [Google Scholar]
- Rabie A.L., Wells J.D., Dent L.K. (1983) The nitrogen content of pollen protein, J. Apicult. Res. 22, 119–123. [Google Scholar]
- Roulston T.H., Buchmann S.L. (2000) A phylogenetic reconsideration of the pollen starch-pollination correlation, Evol. Ecol. Res. 2, 627–643. [Google Scholar]
- Roulston T.H., Cane J.H. (2000) Pollen nutritional content and digestibility for animals, Plant Syst. Evol. 222, 187–209. [Google Scholar]
- Roulston T.H., Cane J.H. (2002) The effect of pollen protein concentration on body size in the sweat bee Lasioglossum zephyrum (Hymenoptera: Apiformes), Evol. Ecol. 16, 49–65. [CrossRef] [Google Scholar]
- Roulston T.H., Cane J.H., Buchmann S.L. (2000) What governs protein content of pollen: pollinator preferences, pollen-pistil interactions, or phylogeny? Ecol. Monogr. 70, 617–643. [Google Scholar]
- Sedivy C., Praz C.J., Müller A., Widmer A., Dorn S. (2008) Patterns of host-plant choice in bees of the genus Chelostoma: the constraint hypothesis of host-range evolution in bees, Evolution 62, 2487–2507. [CrossRef] [PubMed] [Google Scholar]
- Standifer L.N. (1967) A comparison of the protein quality of pollens for growth stimulation and hypopharyngeal glands and longevity of honey bees (Apis mellifera, Hymenoptera, Apidae), Insect. Soc. 14, 415–426. [CrossRef] [Google Scholar]
- Strauss S.Y., Zangerl A.R. (2002) Plant-insect interactions in terrestrial ecosystems, in: Herrera C.M., Pellmyr O. (Eds.), Plant-animal interactions – an evolutionary approach, Blackwell Publishing, Malden, pp. 77–106. [Google Scholar]
- Strickler K. (1979) Specialization and foraging efficiency of solitary bees Hoplitis anthocopoides, Ecology 60, 998–1009. [CrossRef] [Google Scholar]
- Thorp R.W. (1969) Systematics and ecology of bees of the subgenus Diandrena (Hymenoptera: Andrenidae), University of California Publications in Entomology 52, 1–146. [Google Scholar]
- Thorp R.W. (2000) The collection of pollen by bees, Plant Syst. Evol. 222, 211–223. [CrossRef] [Google Scholar]
- Wäckers F.L. (1999) Gustatory response by the hymenopteran parasitoid Cotesia glomerata to a range of nectar and honeydew sugars, J. Chem. Ecol. 25, 2863–2877. [CrossRef] [Google Scholar]
- Westrich P. (1990) Die Wildbienen Baden-Württembergs II, Eugen Ulmer Verlag, Hohenheim. [Google Scholar]
- Wille H., Wille M., Kilchenmann V., Imdorf A., Bühlmann G. (1985) Pollenernte und Massenwechsel von drei Apis mellifera-Völkern auf demselben Bienenstand in zwei aufeinanderfolgenden Jahren, Rev. Suisse Zool. 92, 897–914. [Google Scholar]
- Williams N.M. (2003) Use of novel pollen species by specialist and generalist solitary bees (Hymenoptera: Megachilidae), Oecologia 134, 228–237. [PubMed] [Google Scholar]
- Wisskirchen R., Haeupler H. (1998) Standardliste der Farn- und Blütenpflanzen Deutschlands, Verlag Eugen Ulmer, Stuttgart. [Google Scholar]
All Tables
Analysed plant taxa, their assignment to one of the two tested groups (O = pollen hosts of oligolectic bees, N = not hosting oligolectic bees) and the total concentration of free and protein-bound amino acids as well as percentage of essential amino acids (AA = amino acids).
All Figures
 |
Figure 1 Amino acid profile of two exemplary plant species: Campanula trachelium and Agrimonia eupatoria. C. trachelium hosts oligolectic bees unlike A. eupatoria. All measured amino acids and their derivates are displayed, separated into free and protein-bound fractions. (Arg = arginine, His = histidine, Ile = isoleucine, Leu = leucine, Lys = lysine, Met = methionine, Phe = phenylalanine, Thr = threonine, Trp = tryptophan, Val = valine, α-AAA = α-aminoadipic acid, α-ABA = α-aminobutyric acid, Ala = alanine, Asn = asparagine, Asp = aspartic acid, β-AIBA = β-aminoisobutyric acid, β-Ala = β-Alanine, Car = carnosine, Citr = citrulline, Cys = cysteine, Cyst = cystathionine, GABA = γ-aminobutyric acid, Gln = glutamine, Glu = glutamic acid, Gly = Glycin, 1-Meth = 1-methylhistidine, 3-Meth = 3-methylhistidine, OH-Pro = hydroxyproline, Orn = ornithine, P-Eta = phosphoethanolamine, Pro = proline, P-Ser = phosphoserine, Ser = serine, Tau = taurine, Tyr = tyrosine). |
| In the text | |
 |
Figure 2 Taxonomic signals in pollen amino acids: closely related species often have a similar chemistry. Each symbol denotes one plant species. Plants hosting oligoleges are displayed with « + » plants not known to host them with « o ». Species that plot together are similar in their relative proportions of amino acids (free and protein-bound pooled). Four examples of plant families are highlighted to indicate their similar pollen composition (NMDS, stress = 9.53, Bray-Curtis similarity). |
| In the text | |
 |
Figure 3 Amino acids (AA) compared between plants hosting oligolectic bees vs. plants not
known to host oligoleges. Box whisker plots showing median, quartiles and range.
Plants hosting oligoleges showed a significantly lower pollen quality in terms of
total amino acid concentration (Mann-Whitney U-test; Z = − 2.69, P =
0.0072) and total essential amino acids (Z = − 2.64, P = 0.0082),
whereas the fractions of free and free essential amino acids did not show significant
differences (all Z ≤ 0.92, all |
| In the text | |