| Issue |
Apidologie
Volume 41, Number 2, March-April 2010
|
|
|---|---|---|
| Page(s) | 216 - 224 | |
| DOI | https://doi.org/10.1051/apido/2009079 | |
| Published online | 03 December 2009 | |
Original article
Inferences of evolutionary and ecological events that influenced the population structure of Plebeia remota, a stingless bee from Brazil*
Caractérisation de la structure génétique actuelle des populations de Plebeia remota, abeille sans aiguillon du Brésil, en relation avec les processus de l’évolution et de l’écologie
Populationsgenetik von Plebeia remota, einer stachellosen Biene aus Brasilien, und Rückschlüsse auf evolutionäre und ökologische Ereignisse
Departamento de Genética e Biologia Evolutiva, Instituto de Biociências,
Universidade de São Paulo, Rua do
Matão 277, 05508-090, São
Paulo, SP,
Brazil
Corresponding author: F. de O. Francisco,
fofrancisco@usp.br, mcarias@ib.usp.br
Received:
6
March
2009
Revised:
17
September
2009
Accepted:
1
October
2009
The present study characterised the population genetic structure of Plebeia remota through mitochondrial DNA (mtDNA) analysis and evaluated evolutionary and ecological processes that may have contributed to the species current genetic scenario. Seventy feral nests were sampled representing four geographic regions (Cunha, Curitiba, Prudentópolis, and Blumenau). Fifteen composite mtDNA haplotypes were determined and a high genetic structure was detected among all populations. The current population structure may be a result of queen philopatry and vegetation shifts caused by palaeoclimatic changes and uplift of Brazilian coastal ranges. Finally, this study strongly suggests a revision of the taxonomic status of P. remota from the Prudentópolis region.
Zusammenfassung
Neuere Studien haben gezeigt, dass innerhalb der Art Plebeia remota in ihrem Verbreitungsgebiet ausgeprägte Unterschiede in Verhalten, Morphometrie, chemischen Komponenten und genetischen Besonderheiten existieren. Die genetische Struktur von P. remota wurde mit Hilfe von mitochondrialer DNA (mtDNA) untersucht; evolutionäre und ökologische Prozesse, die zu diesem Szenarium geführt haben könnten wurden abgeleitet. Die Proben wurden in Cunha (São Paulo), Blumenau (Santa Catarina), sowie in Curitiba und Prudentópolis (Paraná) gesammelt (Abb. 1). Die Proben stammen von siebzig Nestern und wurden mittels RFLP Analyse untersucht, wobei 15 Restriktionsenzyme eingesetzt wurden. Die Länge des mtDNA-Moleküls betrug in allen Proben 18 500 bp. Mit zwei der Restriktionsenzyme konnten in keiner der Proben eine Schnittstelle nachgewiesen werden. Mit den restlichen 13 Enzymen wurden insgesamt 27 verschiedene Schnittstellen gefunden; daraus ergaben sich 15 Haplotypen, deren Verteilung gut mit der geographischen Herkunft der Proben korrelierte (Abb. 2). Dabei trat der gleiche Haplotyp nur selten in mehreren Populationen auf (Tab. I). Die Diversität der Haplotypen (h) und der Nukleotide (π) innerhalb jeder einzelnen Population waren vergleichsweise hoch, mit der Ausnahme von Prudentópolis (Tab. II). Die Werte für FST und die Nukleotid-Divergenz zwischen den Populationen sind in Tabelle III dargestellt. Die Analyse der mtDNA zeigte große genetische Variabilität und Isolation zwischen den Populationen, was sowohl durch die Philopatrie der Königinnen als auch mit Habitatfragmentation in früheren Zeiten durch Klimaänderungen oder die Auffaltung des Küstengebirges erklärt werden kann. Zusammen mit bereits veröffentlichten Daten unterstützen die Ergebnisse die Vermutung, dass diese spezifische Population als eine eigene Art angesehen werden sollte. Die Untersuchung zusätzlicher Bienenarten und anderer Organismen aus denselben geographischen Gebieten wird unser Wissen über die Verbreitungsmuster der Arten und die möglichen Zusammenhänge mit historischen und evolutionären Ereignissen in dieser Region vermehren.
Key words: stingless bees / mtDNA / rflp / Meliponini / population genetics
Mots clés : abeille sans aiguillon / ADN mitochondrial / Meliponini / génétique des populations / évolution
Schlüsselwörter: Stachellose Bienen / mtDNA / RFLP / Meliponini / Populationsgenetik
© INRA/DIB-AGIB/EDP Sciences, 2009
1. INTRODUCTION
The stingless bees (tribe Meliponini) exhibit a wide geographic distribution, as they are found in all tropical and subtropical regions on Earth (Wille, 1979; Silveira et al. , 2002). The biogeographical history of this tribe is a controversial issue, and there is no consensus on whether the tribe originated from the Neotropics or Africa (see Kerr and Maule, 1964; Wille, 1979; and some considerations in Rasmussen and Cameron, 2007). The greatest abundance and diversity of stingless bees are found in the Neotropics (Wille, 1979), with approximately 30 genera and 300 species described (Camargo and Pedro, 1992). These data strongly argue in favour of the Neotropical origin (Camargo and Pedro, 1992).
Studies on the biology and ecology of Meliponini have proven their important role in the maintenance of several ecosystems. The pollination of many floral species from the Brazilian Atlantic forest is strictly dependent on these bees (Kerr et al. , 1996). Recently, Bacelar-Lima et al. (2006) described the importance of stingless bees in scattering seeds of the Amazon forest.
The genus Plebeia Schwarz exhibits Neotropical distribution and, according to Michener (2000), is comprised by three subgenera: Plebeia Schwarz, Scaura Schwarz, and Schwarziana Moure. Bees from the subgenus Plebeia are morphologically characterised by small length (3–6 mm) and the presence of whitish or yellow stripes on the face and thorax (Michener, 2000). This subgenus ranges from Mexico to Argentina and is considered a post-Gondwanan group due to its Neotropical endemism. Approximately 30 species of Plebeia have been described (Michener, 2000). Brazil includes 16 recognised species and an unknown number of undescribed species (Silveira et al. , 2002). South-eastern Brazil has been considered the geographical centre of origin for this subgenus (Camargo and Wittmann, 1989).
The species Plebeia remota (Holmberg, 1903) is widely distributed in southern Brazil, occurring in the states of Minas Gerais (MG), São Paulo (SP), Paraná (PR), Santa Catarina (SC), and Rio Grande do Sul (RS) (Fig. 1A) (Wittmann and Hoffmann, 1990; Silveira et al. , 2002; Mouga, 2004). These bees nidify in tree cavities, and a colony can reach 5,000 individuals (van Benthem et al. , 1995). Natural hives are very difficult to be located; the nest’s entrance is very small and only permits the entrance or exit of one individual at a time. Furthermore, workers are very cryptic and any disturbance outside interrupts foraging activity. Ribeiro et al. (2003) reported differences in nest architecture and fall-winter reproductive diapause between P. remota colonies from two geographically distant populations in Brazil: Cunha (SP state) and Prudentópolis (PR state). Further, Patrício and Imperatriz-Fonseca (2004) found differences in the external morphology of queens’ scutella and composition of Dufour glands secretions. Hilário (2005) detected differences in foragers’ flight activity and nest temperature control. Recently, Francisco et al. (2008) also verified differences through mitochondrial DNA (mtDNA), patterns of wing venation, and cuticular hydrocarbons analyses between these two populations.
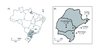 |
Figure 1 (A) Map of Brazil indicating the states of Plebeia remota occurrence. MG: Minas Gerais, SP: São Paulo, PR: Paraná, SC: Santa Catarina, RS: Rio Grande do Sul. (B) Collected sites and number of nests sampled. |
MtDNA is one of the most widely studied molecules for systematics, species characterisation, population structure, and phylogenetic analyses. Analysis by restriction fragment length polymorphism (RFLP) technique allows the investigation of this molecule as a whole, including both conserved and variable regions. Particularly in Meliponini, the mtDNA-RFLP approach has been tested and proved useful for the detection of inter-specific variation within the subgenus Plebeia (Francisco et al. , 2001) and others (Weinlich et al. , 2004; Brito and Arias, 2005; Arias et al. , 2006).
To date, molecular genetic analysis of P. remota has been restricted to mtDNA-RFLP of only few samples (Francisco et al. , 2008); therefore, the available literature for this species is predominantly non-genetic. In this investigation, mtDNA-RFLP analysis was utilised to characterise samples of P. remota collected within its endemic area, representing four distinct populations. Our investigation aimed to: (i) characterise the population structure of P. remota by mtDNA-RFLP analysis, (ii) determine the genetic and evolutionary status of each surveyed population, and (iii) confirm the isolation of the Prudentópolis population. Therefore, this study provides a better understanding of evolutionary and ecological processes that were responsible for the species current population genetic structure.
2. MATERIAL AND METHODS
2.1. Sampling
Adult workers were collected from 70 nests in four localities of Brazil (Fig. 1B): Cunha, SP (n = 18); Prudentópolis, PR (n = 33); Curitiba, PR (n = 7); and Blumenau, SC (n = 12). The samples were initially kept in liquid nitrogen and stored at –80 °C. Bees were checked for species identity by a taxonomist.
2.2. mtDNA analysis
Total DNA was isolated according to the protocol of Sheppard and McPheron (1991), with slight modifications as described in Francisco et al. (2001). The DNA was subjected to single and double digestion overnight with the following 15 restriction enzymes: BamH I, Bcl I,Bgl II, Cfo I, Cla I, EcoR I, EcoR V, Hae III, Hind III, Nde I, Pst I, Pvu II, Sca I, Xba I, and Xho I. The RFLP patterns were verified via Southern blot analysis, as previously described (Francisco et al. , 2001). The PCR-RFLP technique was utilised to identify restriction sites in close proximity. Methods used for PCR reactions, digestions, and visualisations are also described by Francisco et al. (2001).
2.3. Statistical analyses
REAP v4.0 package (McElroy et al. , 1992) was used to calculate the genetic distance between haplotypes (d), nucleotide (π) and haplotype (h) diversities within samples, and nucleotide divergence among populations. The Monte Carlo (Roff and Bentzen, 1989) test was used to determine genetic heterogeneity among populations. ARLEQUIN v3.11 (Excoffier et al. , 2005) was used to detect population differentiation through the Markov chain method and exact test. AMOVA (Excoffier et al. , 1992) was used for calculation of the intra- and inter-population diversity and ΦST and FST values. Based on d values, unrooted minimum evolution (ME) trees (Rzhetsky and Nei, 1992) were estimated using the program MEGA4 (Tamura et al. , 2007). Plebeia saiqui served as outgroup. Mantel tests (10 000 permutations) were performed by GENEPOP package v3.4 (Raymond and Rousset, 1995) to correlate genetic (nucleotide divergence and FST) and geographical distances.
3. RESULTS
MtDNA from 70 nests was screened with 15 restriction endonucleases. Fragment size and number were calculated from Southern blot membranes, and the total length of mtDNA for all samples was estimated to be approximately 18 500 bp. Two restriction enzymes (Cfo I and Pvu II) did not cut the mtDNA for any sample. The remaining 13 enzymes generated a total of 27 different restriction sites, and 15 haplotypes were obtained (see figure in the electronic supplementary material). Most haplotypes were not shared among populations (Tab. I). Only haplotypes h01 and h02 were both present in Cunha and Curitiba populations. The ME tree based on genetic distances between haplotypes (Fig. 2) defined four clusters, which were well related to the respective geographic origin: Prudentópolis, Blumenau, Curitiba, and Cunha. Notably, haplotype h10 (Curitiba) is genetically distant from all haplotypes, but closely related to the outgroup P. saiqui. Haplotype (h) and nucleotide (π) diversities within each population were relatively high, except in Prudentópolis (Tab. II). Samples from Prudentópolis and Curitiba presented the lowest and the highest π values of 0.2191 and 2.2383%, respectively.
 |
Figure 2 Minimum evolution tree of mitochondrial DNA haplotypes based on genetic distance. The species Plebeia saiqui was included as the outgroup. |
Haplotype frequencies and distribution detected in four populations of Plebeia remota.
Haplotype (h) and percentage nucleotide (π) diversity within the populations of Plebeia remota.
Values of FST and nucleotide divergence between populations are shown in Table III. Cunha and Curitiba presented the lowest nucleotide divergence (0.0060), and in contrast, Cunha and Prudentópolis presented the highest (0.0386). High differentiation among the four populations (0.1659 to 0.7266) was observed according to FST values.
The Monte Carlo distribution indicated significant heterogeneity of haplotype frequencies (P < 0.0001) among population pairs. Exact tests, which were also based on haplotype frequencies, exhibited differentiation among population pairs (all P < 0.0114). The analysis of molecular variance (AMOVA) revealed significant genetic variation within and among populations (41.65% and 58.35%, respectively). The ΦST value also revealed a highly significant population structure (ΦST = 0.5835; P < 0.0001). Mantel’s tests based on nucleotide divergence and FST values demonstrated that the genetic distances were not correlated with the geographical distances (P = 0.2942 and P = 0.4184, respectively).
Pairwise comparisons of nucleotide divergence (above diagonal) and FST estimates (below diagonal) between pairs of Plebeia remota populations.
4. DISCUSSION
4.1. Causes of interpopulation mtDNA differentiation
The distribution of mtDNA haplotypes and statistical analyses demonstrated a clear isolation among the four populations, suggesting an absence of gene flow mediated by queens. This result can be explained in part by the swarming behaviour of stingless bees. The new nest is built only a few metres from the mother colony and keeps a high dependence on it until a virgin queen and a group of workers migrate to the new site (Michener, 1979; Engels and Imperatriz-Fonseca, 1990). Queen philopatric behaviour has been associated to population structure in other social insects (Ross, 2001).
Palaeoclimatic and palaeogeographic events should also be considered here since they affected drastically the landscape. It is well known that the climatic changes in Pliocene and Pleistocene led to habitat fragmentation which in turn may have promoted speciation and extinction events, and organism diversity as an ultimate consequence (Potts and Behrensmeyer, 1992). The current distribution of several organisms in Europe has been associated to Pleistocene climatic changes (Hewitt, 1999). South America was not subjected to severe glaciation, but the climate was strongly altered (Hewitt, 1996). Glaciations began approximately 2.5 million years (Myr) before present (BP) in the Late Pliocene (Hewitt, 1996, 1999) and were more intense during the Pleistocene, with cycles of 100 000 yr interrupted by short warm periods (Potts and Behrensmeyer, 1992; Hewitt, 1996). Forests expanded during warm and moist periods and then contracted after the climate became cold and dry. Differentiation among P. remota populations may have occurred during forest contractions. The high rate of exclusive haplotypes and the high values of FST are indicative of an ancient isolation. Considering the rate of 2% divergence per 1 Myr for mtDNA coding sequences (De Salle et al. , 1987), bees from Prudentópolis and Cunha can be inferred to be isolated for 1.9 Myr (Late Pliocene). The population from Prudentópolis seems to have been isolated from the other populations for approximately 1.5–1.4 Myr (Early Pleistocene). Conversely, populations from Cunha and Curitiba presented the most recent isolation of approximately 300 000 yr BP (Late Pleistocene), and the two haplotypes common to both populations (h01 and h02) can be remnants of the haplotype diversity before ancestral population split. Although inferences of evolutionary time based on molecular clocks should be used with caution (Pulquério and Nichols, 2007), other studies have successfully applied such estimates to understand bee population structure dynamics. For example, Franck et al. (2000) suggested that glaciation has isolated lineages of A. mellifera in Italian refuges at approximately 190 000 yr BP. Dick et al. (2004) estimated that the divergence time among cross-Andean Euglossini bees ranges from 0 to 1.42 Myr BP, indicating cross-Andean dispersal after the uplift of the Andes mountain range.
The uplift of coastal ranges (Serra do Mar) also exhibited a strong influence over the flora distribution in southern and south-eastern Brazil and, consequently, might have isolated populations by habitat fragmentation (Lara and Patton, 2000). The uplift began in the Palaeocene, about 65 Myr BP (Petri and Fúlfaro, 1988) and lasted until the Pleistocene (Freitas, 1951). Ancient habitat fragmentation caused by climatic changes, uplift of coastal ranges, and also female philopatry may have been responsible for the isolation detected among P. remota populations.
4.2. Causes of intrapopulation mtDNA diversity
Interestingly, the lowest mtDNA genetic diversity was found in the region with the largest sample size (Prudentópolis, n = 33). Although four haplotypes were found in Prudentópolis, three were very rare, and each was found in only one nest. Haplotype h05 was the predominant form, being present in the remaining 30 nests. Moreover, the haplotypes found in Prudentópolis were highly similar as evidenced by the low π value, which indicates a shorter coalescence time and suggests a recent bottleneck or a small founder population. In a palynological study, Behling (1997) demonstrated that the last cold and dry period in the Paraná highlands was approximately 12 480–9660 14C yr BP. At that time, vegetation was predominantly grassland and the forests were restricted to moist environments such as low-elevation valleys. As Prudentópolis is located in the Paraná highlands, the habitat restrictions abovementioned certainly affected this region and moreover may have confined bees to refuges. After this period, the climate gradually became moister, which allowed the expansion of forests and bee populations. Our data suggest that habitat re-colonisation was performed mainly by bees carrying haplotype h05, and the other three haplotypes may have appeared in the populations as a result of new mutations.
In contrast, samples from Curitiba presented higher haplotype and nucleotide diversities. Unexpectedly, these values were obtained from a low sample size (7 nests). These results indicate that Curitiba may represent an ancient population, and that this region is the possible centre of origin for the species. Similar results obtained for A. mellifera strongly support the northeastern Africa as the species origin centre, since three mitochondrial lineages exist simultaneously in that region (Franck et al. , 2001). The high genetic variability verified in the population from Curitiba may be interpreted as a result from the climatic changes in the Late Pleistocene and Holocene, leading to the existence of several refuges in this geographic area. Indeed, Curitiba is in close proximity to Serra do Mar, a mountain chain that follows the Brazilian coast from Santa Catarina to Espírito Santo states. During the cold and dry period, the Atlantic forest lost continuity and was restricted to refuges with high humidity and substantial orographic rain (Ab’Saber, 1977, 1979) in the Atlantic Ocean slope. Therefore, multiple refuges would have allowed the evolution of multiple haplotypes.
Palynological evidence of contraction and expansion of forests in the Late Pleistocene and Holocene has also been found in regions near Cunha (Behling et al. , 2007) and Blumenau (Behling and Negrelle, 2001), indicating that the intrapopulational diversity of P. remota in those regions may also be explained by these events.
5. CONCLUSIONS
This work demonstrated that the four studied populations exhibited exclusive genetic characteristics. The population from Prudentópolis is the most genetically distant and may have diverged from the ancestral populations in the Late Pliocene. Previous data had already reported unique ecologic, morphologic, and chemical characteristics of this population (Francisco et al. , 2008). Such evidences support the hypothesis that it constitutes a new species. Indeed, when nests from Cunha and Prudentópolis were kept in a meliponary for five years, hybrids were not observed (molecular data not shown).
It is worth to note the genetic distance observed for haplotype h10 in relation to all samples. This haplotype was found in one nest from Curitiba. The addition of more molecular, morphological, and ecological data from a larger sample size from Curitiba region will certainly improve our understanding of the taxonomic status of this particular sample.
The study of additional bee species and also other organisms from the same geographic areas analyzed here will be very important to validate or not the hypotheses we have raised and certainly will increase our knowledge about the species distribution pattern and the possible association with historic and evolutionary events that occurred in those regions.
Online material
 |
Supplementary Figure 1. Linearised restriction site maps and gene positions of the 15 haplotypes of Plebeia remota mitochondrial genomes. M: BamH I, B: Bcl I, G: Bgl II, C: Cla I, E: EcoR I, V: EcoR V, A: Hae III, D: Hind III, N: Nde I, P: Pst I, S: Sca I, X: Xba I, and H: Xho I. ND5: NADH dehydrogenase subunit 5, ND4: NADH dehydrogenase subunit 4, ND6: NADH dehydrogenase subunit 6, cytB: cytochrome B, ND1: NADH dehydrogenase subunit 1, 16S: large subunit ribosomal RNA, 12S: small subunit ribosomal RNA, ND2: NADH dehydrogenase subunit 2, COI: cytochrome c oxidase subunit 1, COII: cytochrome c oxidase subunit 2, 8: ATP FO synthase subunit 8, 6: ATP FO synthase subunit 6, COIII: cytochrome c oxidase subunit 3, and ND3: NADH dehydrogenase subunit 3. |
Acknowledgments
We thank V.L. Imperatriz-Fonseca, M.F. Ribeiro, G. Moretto, L. Muegge, S. Gonzaga, and C. Chociai for samples, S. Coelho for technical support, G.A.R. Melo for morphological identification, R.M. Brito and the two anonymous reviewers for their comments and suggestions on an earlier version of this report. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 99/11190-6).
References
- Ab’Saber A.N. (1977) Espaços ocupados pela expansão dos climas secos da América do Sul, por ocasião dos períodos glaciais quaternários, Paleoclimas 3, 1–19. [Google Scholar]
- Ab’Saber A.N. (1979) Os mecanismos da desintegração das paisagens tropicais no Pleistoceno: efeitos paleoclimáticos do período Würm-Winsconsin no Brasil, Paleoclimas 8, 1–11. [Google Scholar]
- Arias M.C., Brito R.M., Francisco F.O., Moretto G., Oliveira F.F., Silvestre D., Sheppard W.S. (2006) Molecular markers as a tool for population and evolutionary studies of stingless bees, Apidologie 37, 259–274. [CrossRef] [EDP Sciences] [Google Scholar]
- Bacelar-Lima C.G., Freire D.C.B., Coletto-Silva A., Costa K.B., Laray J.P.B., Vilas-Boas H.C., Carvalho-Zilse G.A. (2006) Melitocoria de Zygia racemosa (Ducke) Barneby and Grimes por Melipona seminigra merrillae Cockerell, 1919 y Melipona compressipes manaosensis Schwarz, 1932 (Hymenoptera, Meliponina) en la Amazonía Central, Brasil, Acta Amaz. 36, 343–348. [Google Scholar]
- Behling H. (1997) Late Quaternary vegetation, climate and fire history of the Araucaria forest and campos region from Serra Campos Gerais, Paraná State (South Brazil), Rev. Palaeobot. Palyno. 97, 109–121. [CrossRef] [Google Scholar]
- Behling H., Negrelle R.R.B. (2001) Tropical rain forest and climate dynamics of the Atlantic lowland, Southern Brazil, during the Late Quaternary, Quaternary Res. 56, 383–389. [CrossRef] [Google Scholar]
- Behling H., Dupont L., Safford H.D., Wefer G. (2007) Late Quaternary vegetation and climate dynamics in the Serra da Bocaina, southeastern Brazil, Quatern. Int. 161, 22–31. [CrossRef] [Google Scholar]
- Brito R.M., Arias M.C. (2005) Mitochondrial DNA characterization of two Partamona species (Hymenoptera, Apidae, Meliponini) by PCR+RFLP and sequencing, Apidologie 36, 431–438. [CrossRef] [EDP Sciences] [Google Scholar]
- Camargo J.M.F., Pedro S.R.M. (1992) Systematics, phylogeny and biogeography of the Meliponinae (Hymenoptera, Apidae): a mini review, Apidologie 23, 509–522. [CrossRef] [EDP Sciences] [Google Scholar]
- Camargo J.M.F., Wittmann D. (1989) Nest architecture and distribution of the primitive stingless bee, Mourella caerulea (Hymenoptera, Apidae, Meliponinae): evidence for the origin of Plebeia (s. lat.) on the Gondwana Continent, Stud. Neotrop. Fauna E. 24, 213–229. [CrossRef] [Google Scholar]
- De Salle R., Freedman T., Prager E.M., Wilson, A.C. (1987) Tempo and mode of sequence evolution in mitochondrial DNA of Hawaiian Drosophila, J. Mol. Evol. 26, 157–164. [CrossRef] [PubMed] [Google Scholar]
- Dick C.W., Roubik D.W., Gruber K.F., Bermingham E. (2004) Long-distance gene flow and cross-Andean dispersal of lowland rainforest bees (Apidae: Euglossini) revealed by comparative mitochondrial DNA phylogeography, Mol. Ecol. 13, 3775–3785. [CrossRef] [PubMed] [Google Scholar]
- Engels W., Imperatriz-Fonseca V.L. (1990) Caste development, reproductive strategies and control of fertility in honeybees and stingless bees, in: Engels W. (Ed.), Social Insects: An Evolutionary Approach to Castes and Reproduction, Springer-Verlag, Berlin, pp. 166–230. [Google Scholar]
- Excoffier L., Laval G., Schneider S. (2005) Arlequin ver. 3.0: An integrated software package for population genetics data analysis, Evol. Bioinf. Online 1, 47–50. [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data, Genetics 131, 479–491. [PubMed] [Google Scholar]
- Francisco F.O., Nunes-Silva P., Francoy T.M., Wittmann D., Imperatriz-Fonseca V.L., Arias M.C., Morgan E.D. (2008) Morphometrical, biochemical and molecular tools for accessing biodiversity. An example in Plebeia remota, Insect. Soc. 55, 231–237. [CrossRef] [Google Scholar]
- Francisco F.O., Silvestre D., Arias M.C. (2001) Mitochondrial DNA characterization of five species of Plebeia (Apidae: Meliponini): RFLP and restriction maps, Apidologie 32, 323–332. [CrossRef] [EDP Sciences] [Google Scholar]
- Franck P., Garnery L., Celebrano G., Solignac M., Cornuet, J.-M. (2000) Hybrid origins of honeybees from Italy (Apis mellifera ligustica) and Sicily (A. m. sicula), Mol. Ecol. 9, 907–921. [CrossRef] [PubMed] [Google Scholar]
- Franck P., Garnery L., Loiseau A., Oldroyd B.P., Hepburn H.R., Solignac M., Cornuet, J.-M. (2001) Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data, Heredity 86, 420–430. [CrossRef] [PubMed] [Google Scholar]
- Freitas R.O. (1951) Ensaio sobre o relevo tectônico do Brasil, Rev. Bras. Geogr. 2, 171–222. [Google Scholar]
- Hewitt G.M. (1996) Some genetic consequences of ice ages, and their role in divergence and speciation, Biol. J. Linn. Soc. 58, 247–276. [Google Scholar]
- Hewitt G.M. (1999) Post-glacial re-colonization of European biota, Biol. J. Linn. Soc. 68, 87–112. [Google Scholar]
- Hilário S.D. (2005) Atividade de vôo e termorregulação de Plebeia remota (Holmberg, 1903) (Hymenoptera, Apidae, Meliponini), PhD thesis, Universidade de São Paulo. [Google Scholar]
- Kerr W.E., Maule V. (1964) Geographic distribution of stingless bees and its implications (Hymenoptera: Apidae), J. New York Entomol. S. 72, 2–18. [Google Scholar]
- Kerr W.E., Carvalho G.A., Nascimento V.A. (1996) Abelha Uruçu: Biologia, Manejo e Conservação, Fundação Acangaú, Belo Horizonte. [Google Scholar]
- Lara M.C., Patton J.L. (2000) Evolutionary diversification of spiny rats (genus Trinomys, Rodentia: Echimyidae) in the Atlantic Forest of Brazil, Zool. J. Linn. Soc. Lond. 130, 661–686. [CrossRef] [Google Scholar]
- McElroy D., Moran P., Bermingham E., Kornfield I. (1992) REAP: an integrated environment for the manipulation and phylogenetic analysis of restriction data, J. Hered. 83, 157–158. [PubMed] [Google Scholar]
- Michener C.D. (1979) Biogeography of the bees, Ann. Mo. Bot. Gard. 66, 277–347. [CrossRef] [Google Scholar]
- Michener C.D. (2000) The Bees of the World, The John Hopkins University Press, Baltimore. [Google Scholar]
- Mouga S.M.D.S. (2004) As comunidades de abelhas (Hymenoptera, Apoidea) em Mata Atlântica na região nordeste do Estado de Santa Catarina, Brasil, PhD thesis, Universidade de São Paulo. [Google Scholar]
- Patrício E.F.R.A., Imperatriz-Fonseca V.L. (2004) S.E.M. and GC-MS studies of Plebeia remota (Hymenoptera, Meliponini), Proceedings of the 8th IBRA International Conference on Tropical Bees and IV Encontro sobre Abelhas, p. 739. [Google Scholar]
- Petri S., Fúlfaro V.J. (1988) Geologia do Brasil (Fanerozóico), EDUSP, São Paulo. [Google Scholar]
- Potts R., Behrensmeyer A.K. (1992) Late Cenozoic terrestrial ecosystems, in: Behrensmeyer A.K., Damuth J.D., DiMichele W.A., Potts R. (Eds.), Terrestrial Ecosystems Through Time: Evolutionary Paleoecology of Terrestrial Plants and Animals, Chicago University Press, Chicago, pp. 419–541. [Google Scholar]
- Pulquério M.J.F., Nichols R.A. (2007) Dates from the molecular clock: how wrong can we be? Trends Ecol. Evol. 22, 180–184. [CrossRef] [PubMed] [Google Scholar]
- Rasmussen C., Cameron S.A. (2007) A molecular phylogeny of the Old World stingless bees (Hymenoptera: Apidae: Meliponini) and the non-monophyly of the large genus Trigona, Syst. Entomol. 32, 26–39. [CrossRef] [Google Scholar]
- Raymond M., Rousset F. (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism, J. Hered. 86, 248–249. [Google Scholar]
- Ribeiro M.F., Imperatriz-Fonseca V.L., Santos-Filho P.S. (2003) A interrupção da construção de células de cria e postura em Plebeia remota (Holmberg) (Hymenoptera, Apidae, Meliponini), in: Melo G.A.R., Alves-dos-Santos I. (Eds.), Apoidea Neotropica: Homenagem aos 90 Anos de Jesus Santiago Moure, Editora da UNESC, Criciúma, pp. 177–188. [Google Scholar]
- Roff D.A., Bentzen P. (1989) The statistical analysis of mitochondrial DNA polymorphisms: χ2 and the problem of small samples, Mol. Biol. Evol. 6, 539–545. [PubMed] [Google Scholar]
- Ross K.G. (2001) Molecular ecology of social behaviour: analyses of breeding systems and genetic structure, Mol. Ecol. 10, 265–284. [CrossRef] [PubMed] [Google Scholar]
- Rzhetsky A., Nei M. (1992) A simple method for estimating and testing minimum evolution trees, Mol. Biol. Evol. 9, 945–967. [Google Scholar]
- Sheppard W.S., McPheron B.A. (1991) Ribosomal DNA diversity in Apidae, in: Smith D.R. (Ed.), Diversity in the Genus Apis, Westview Press, Boulder, pp. 89–102. [Google Scholar]
- Silveira F.A., Melo G.A.R., Almeida E.A.B. (2002) Abelhas Brasileiras: Sistemática e Evolução, F.A. Silveira, Belo Horizonte. [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0, Mol. Biol. Evol. 24, 1596–1599. [Google Scholar]
- van Benthem F.D.J., Imperatriz-Fonseca V.L., Velthuis, H.H.W. (1995) Biology of the stingless bee Plebeia remota (Holmberg): observations and evolutionary implications, Insect. Soc. 42, 71–87. [CrossRef] [Google Scholar]
- Weinlich R., Francisco F.O., Arias M.C. (2004) Mitochondrial DNA restriction and genomic maps of seven species of Melipona (Apidae: Meliponini), Apidologie 35, 365–370. [CrossRef] [EDP Sciences] [Google Scholar]
- Wille A. (1979) Phylogeny and relationships among the genera and subgenera of the stingless bees (Meliponinae) of the world, Rev. Biol. Trop. 27, 241–277. [Google Scholar]
- Wittmann W., Hoffman M. (1990) Bees of Rio Grande do Sul, Southern Brazil, Iheringia, Sér. Zool. 70, 17–43. [Google Scholar]
All Tables
Haplotype frequencies and distribution detected in four populations of Plebeia remota.
Haplotype (h) and percentage nucleotide (π) diversity within the populations of Plebeia remota.
Pairwise comparisons of nucleotide divergence (above diagonal) and FST estimates (below diagonal) between pairs of Plebeia remota populations.
All Figures
 |
Figure 1 (A) Map of Brazil indicating the states of Plebeia remota occurrence. MG: Minas Gerais, SP: São Paulo, PR: Paraná, SC: Santa Catarina, RS: Rio Grande do Sul. (B) Collected sites and number of nests sampled. |
| In the text | |
 |
Figure 2 Minimum evolution tree of mitochondrial DNA haplotypes based on genetic distance. The species Plebeia saiqui was included as the outgroup. |
| In the text | |
 |
Supplementary Figure 1. Linearised restriction site maps and gene positions of the 15 haplotypes of Plebeia remota mitochondrial genomes. M: BamH I, B: Bcl I, G: Bgl II, C: Cla I, E: EcoR I, V: EcoR V, A: Hae III, D: Hind III, N: Nde I, P: Pst I, S: Sca I, X: Xba I, and H: Xho I. ND5: NADH dehydrogenase subunit 5, ND4: NADH dehydrogenase subunit 4, ND6: NADH dehydrogenase subunit 6, cytB: cytochrome B, ND1: NADH dehydrogenase subunit 1, 16S: large subunit ribosomal RNA, 12S: small subunit ribosomal RNA, ND2: NADH dehydrogenase subunit 2, COI: cytochrome c oxidase subunit 1, COII: cytochrome c oxidase subunit 2, 8: ATP FO synthase subunit 8, 6: ATP FO synthase subunit 6, COIII: cytochrome c oxidase subunit 3, and ND3: NADH dehydrogenase subunit 3. |
| In the text | |


