| Issue |
Apidologie
Volume 41, Number 2, March-April 2010
|
|
|---|---|---|
| Page(s) | 194 - 202 | |
| DOI | https://doi.org/10.1051/apido/2009075 | |
| Published online | 26 January 2010 | |
Original article
Reduced expression of major royal jelly protein 1 gene in the mushroom bodies of worker honeybees with reduced learning ability*
Expression réduite du gène de la protéine majeure 1 de la gelée royale dans les corps pédonculés des ouvrières d’abeille avec diminution de la capacité d’apprentissage
Verringerte Expression des Gelee Royal Protein 1-Gens in den Pilzkörpern von Arbeiterinnen der Honigbiene nach Isolation vom Volk
1
Faculty of Agriculture, Tamagawa University, 6-1-1 Tamagawagakuen, Machida, Tokyo
194-8610,
Japan
2
Honeybee Science Research Center, Tamagawa University,
6-1-1 Tamagawagakuen, Machida
Tokyo
194-8610,
Japan
Corresponding author: M. Hojo, hojoj@tmu.ac.jp
Received:
25
April
2009
Revised:
19
August
2009
Accepted:
4
September
2009
The learning ability of European honeybees, Apis mellifera, develops with age. However, when worker bees are isolated from their colony and are fed only sucrose solution, their learning development is hindered. This rearing method has allowed us to compare worker bees of the same age but with different learning abilities. In this study, we examined the influence of this rearing condition on gene expression in the mushroom body, which is the insect brain center involved in learning and memory. A differential display experiment comparing worker bees maintained in a hive with those reared in isolation showed that the expression of the major royal jelly protein (mrjp) 1 gene was reduced in the isolated worker bees. MRJP1 is synthesized in the hypopharyngeal gland and serves a nutritional function in larval and queen food. Our results suggest that mrjp1 is also important in brain function, possibly involved in the development of learning ability.
Zusammenfassung
Die Lernfähigkeiten der Westlichen Honigbiene, Apis mellifera, entwickeln sich mit zunehmendem Alter der Arbeiterin weiter. Das Duftlernen ist ein Beispiel hierfür. Wenn junge Arbeiterinnen allerdings in Isolation ohne ihr Volk gehalten werden und ausschließlich mit Zuckerlösung gefüttert werden, wird ihre Lernentwicklung behindert. Diese Aufzuchtmethode erlaubte uns, Arbeiterinnen desselben Alters mit unterschiedlichen Lernfähigkeiten zu untersuchen. Während unserer Studie untersuchten wir den Einfluss der Aufzuchtbedingung auf die Genexpression im Pilzkörper, eines Bereiches im Insektengehirn, dem eine zentrale Rolle beim Lernen und Gedächtnis zukommt. Ein differential display Experiment, bei dem Bienen aus dem Volk mit isoliert aufgezogenen Arbeiterinnen verglichen wurden, zeigt, dass die Expression des major royal jelly protein (mrjp1) in isolierten Bienen deutlich reduziert ist. Frisch geschlüpfte Bienen wurden hierzu einzeln 8 Tage lang bei 32 °C im Dunkeln in Glasfläschchen gehalten und mit Zuckerwasser gefüttert. Mithilfe der Fluorescence differential display (FDD) Technik mit 16 primer sets wurde die Genexpression im Pilzkörper gemessen. Von insgesamt 604 Banden haben wir 13 Banden gefunden, die bei sozial aufgewachsenen Bienen eine höhere Expressionsintensität aufwiesen als in den isolierten Bienen. Eine Bande von etwa 1,1 kbp identifizierten wir als mrjp1. Diesen Befund konnten wir mit einer quantitativen real-time PCR bestätigen. Unsere Ergebnisse deuten an, dass mrjp1 wichtig für die Gehirnfunktion ist und möglicherweise an der Entwicklung von Lernfähigkeiten beteiligt ist.
Key words: Apis mellifera / mrjp / gene expression / differential display / social environment
Mots clés : Apis mellifera / protéine majeure de la gelée royale / expression génique / apprentissage / environnement social
Schlüsselwörter: Apis mellifera / Genexpression / Lernfähigkeiten / Soziale Umgebung
© INRA/DIB-AGIB/EDP Sciences, 2009
1. INTRODUCTION
The honeybee Apis mellifera is a model insect for molecular studies of social behaviour (Robinson et al. , 1997, 2005; Evans and Wheeler, 2001; Whitfield et al. , 2002, 2003, 2006; Cash et al. , 2005; The Honey Bee Genome Sequencing Consortium, 2006; Barchuk et al. , 2007). Worker honeybees change their tasks depending on age (age polyethism): young workers feed larvae in the hive (nursing) and older workers, 2–3-week-old after adult emergence, work outside the hive to collect nectar and pollen (foraging) (Robinson, 1992). The foragers search a wide area around the hive for food sources, and share information about preferable foraging sites with the nest mates through dance communication (Von Frisch, 1967; Winston, 1987; Menzel and Mueller, 1996). Because of such behavioural complexity, foragers are assumed to have a greater information processing ability compared to the nurses.
In honeybees, learning and memory, especially olfactory learning, are thought to be formed through integration of sensory information in the mushroom bodies (Erber et al. , 1980; Durst et al. , 1994; Fahrbach and Robinson, 1995; Hammer and Menzel, 1995; Meller and Davis, 1996; Rybak and Menzel, 1998; Heisenberg, 1998; Menzel, 2001; Menzel and Giurfa, 2001). Gene expression in mushroom bodies has been examined to identify genes that are involved in learning and memory in honeybees, and some genes are found to be specifically expressed in the mushroom body intrinsic neurons known as Kenyon cells (Kamikouchi et al. , 1998, 2000; Takeuchi et al. , 2001, 2002, 2004; Sawata et al. , 2002; Yamazaki et al. , 2006).
Ichikawa and Sasaki (2003) examined olfactory associative learning using a proboscis extension conditioning assay (Bitterman et al. , 1983) and reported that learning ability of workers develops with age, but this development is severely reduced when they are reared in isolation and provided with sucrose as the only food. The method of Ichikawa and Sasaki (2003) provides an experimental system to compare the gene expression profiles of same age workers with different learning abilities. In the present study, we compared the gene expression profiles in the mushroom bodies of workers reared in a hive and those reared by the method of Ichikawa and Sasaki (2003) by differential display to identify the genes involved in the development of learning ability in honeybees. We report that the major royal jelly protein (mrjp) 1 gene, which is expressed in the hypopharyngeal gland, is also expressed in the mushroom bodies, and its expression is reduced when worker bees are reared in isolation. We also report on the expression of the other mrjp genes (mrjp2–9) in the mushroom bodies of workers.
2. MATERIALS AND METHODS
2.1. Rearing of workers in isolation
European honeybees, A. mellifera, were purchased from a local supplier (Nonogaki Apiary, Aichi, Japan) and maintained on the bee farm of Tamagawa University, Machida, Japan. Workers were kept in an isolated condition as reported previously by Ichikawa and Sasaki (2003). In brief, newly emerged workers younger than 1 day were individually confined to a glass vial (3 cm in diameter, 5 cm in high) containing another small glass vial (ø1 cm, 3 cm high) with 1 M sucrose. The vials were covered with clean cheesecloth and buried in charcoal in a plastic case. The sucrose solution was replaced once every three days. These workers, referred to as ‘isolated bees’, were reared in a dark incubator at 32 °C for eight days.
To obtain known-age workers from a hive, newly emerged workers were marked with colour paint and returned to their hive. The marked workers that were maintained in the hive for 8 days were referred to as ‘colony bees’.
2.2. Fluorescent differential display
The central region of the worker brain, including the mushroom bodies, was dissected out in RNAlater RNA stabilization reagent (Qiagen, Tokyo, Japan). Antennal lobes and optic lobes were carefully removed as described by Takeuchi et al. (2001). The total RNA was extracted from mushroom bodies of 8 to 16 individuals using the RNeasy mini kit (Qiagen), and the RNA concentration was determined by a spectrophotometer (GeneQuant pro; GE Healthcare Bio-Sciences, Tokyo, Japan).
To perform fluorescence differential display (FDD), 1 μg of RNA was first treated with 1 unit of RNase-free DNase I (Invitrogen Corp., Carlsbad, CA, USA). FDD reactions were performed using the fluorescence differential display kit (Takara Bio Inc. Otsu, Shiga, Japan) according to the manufacturer’s instructions. In brief, the DNase-treated RNA was reverse transcribed with 25 units of AMV reverse transcriptase XL (Takara Bio Inc., Otsu, Shiga, Japan) with 275 pmol of rhodamine-labelled primer (5’-T13−15GC-3’). The polymerase chain reaction (PCR) was performed with upstream primers (No. 9 to 24) and a rhodamine-labelled downstream primer. The PCR products were separated on a 6% polyacrylamide denaturing gel and the gel was scanned using a fluorescent image analyzer (FMBIO-III Multi-View; Hitachi Software Engineering, Tokyo, Japan). Reactions without reverse transcriptase were used as negative controls to confirm that the RNA preparation was not contaminated with genomic DNA. The FDD-PCR reactions were performed in duplicate using different sets of RNA samples to examine the reproducibility of the analysis.
2.3. Reamplification, subcloningand sequencing
Bands of interest were excised, and the gel was boiled in deionized water to extract DNA. The extracted DNA was reamplified by PCR using TaKaRa LA Taq (Takara Bio) with the same primer combination used in the FDD, but without the fluorescent label in the downstream primer under the following conditions: 94 °C for 2 min, 40 °C for 5 min and 72 °C for 5 min for 1 cycle; 94 °C for 30 s, 40 °C for 2 min and 72 °C for 1 min for 35 cycles and then 72 °C for 5 min. The PCR product was purified with the QIAEX II gel extraction kit (Qiagen), ligated into pGEM-T easy vector (Promega, Madison, WI, USA) and transfected into Escherichia coli JM109 competent cells (Takara Bio). The nucleotide sequence of the inserted DNA fragment was determined using the BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster, CA, USA) and the ABI PRISM 3100 genetic analyzer (Applied Biosystems). The identified sequence was subjected to a homology search using the basic local alignment search tool (BLAST) on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov/BLAST/).
2.4. Real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNA was extracted from mushroom bodies using the RNeasy Mini kit (Qiagen). DNase I treatment was included in the RNA extraction protocol according to the manufacturer’s instruction. The RNA was then reverse transcribed using TaqMan reverse transcription reagents (Applied Biosystems) with oligo(dT) primer.
The qRT-PCR was performed using a SYBR Green I chemistry and sequence detection system ABI PRISM 7000 (Applied Biosystems). Primers specific to the target genes were designed using primer express software (Applied Biosystems). The optimal primer concentrations for PCR were determined for each primer set by comparing amplification profiles with different primer concentrations. The primer sequences and primer concentrations employed are shown in Table I. Amplification was performed in a 25-μL reaction volume containing 12.5 μL of Power SYBR Green PCR master mix (Applied Biosystems). The thermal cycling program consisted of 2 min at 50 °C and 10 min at 95 °C followed by 50 cycles of 15 s at 95 °C and 1 min at 60 °C. To confirm specific amplification of each cDNA fragment, the PCR products were electrophoresed on an agarose gel on which a single band of the expected size was observed for each gene. The PCR products were also sequenced to confirm their identities.
Primer sets of major royal jelly protein (mrjp) genes for real-time quantitative reverse transcriptase polymerase chain reaction.
Each cDNA sample was measured in triplicate, and the levels of target transcripts were normalized to actin (accession number AB023025), which is a house keeping gene and expressed equally in different honeybee castes (Chen et al. , 2005). The data were analyzed by the relative standard curve method according to the user’s manual (Applied Biosystems, 2001).
3. RESULTS
3.1. Comparison of gene expression profiles by differential display
We compared gene expression in the mushroom bodies of worker bees maintained in a colony (colony bees) with those reared in isolation (isolated bees) by fluorescence differential display (FDD). Using 16 primer sets, we detected a total of 604 bands from the colony bees and 594 bands from the isolated bees. Of these, 591 bands showed almost equal expression intensity between the colony and isolated bees, 13 bands had stronger expression in the colony bees than in the isolated bees and three bands had weaker expression in the colony bees than in the isolated bees. Although differential display has low reproducibility (Livesey and Hunt, 1996), our results were reproducible in two independent runs with different RNA samples. Of the colony bee-specific bands, a band of approximately 1.1 kbp using No. 13 upstream primer showed the most obvious difference in intensity (Fig. 1), and we determined the nucleotide sequence of this band. A BLAST search showed that the 1047-bp sequence was identical to that of major royal jelly protein (mrjp) 1 (accession No. AF000633).
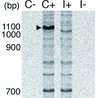 |
Figure 1 An electrophoresis profile of the fluorescent differential display comparing gene expression in the mushroom bodies of colony bees (C+) and isolated bees (I+). Numbers indicate molecular size (bp). Arrowhead indicates approximately 1.1-kbp band of the colony bee-specific gene. No bands were detected in the negative (RT-) control lanes (C- and I-). |
3.2. Quantification of mrjp1 expression by real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
To further confirm the differential expression of mrjp1 identified in the differential display analysis, we performed qRT-PCR. Each RNA sample was extracted from a pool of 20 mushroom bodies from colony and isolated bees. The measurements were performed using three pairs of RNA samples extracted at three different times. The results showed that mrjp1 expression was clearly higher in the colony bees than in the isolated bees. The expression level of mrjp1 in the isolated bees was less than 1% of those in the colony bees in the three comparisons (Fig. 2).
 |
Figure 2 Relative major royal jelly protein (mrjp) 1 gene expression level measured by real-time quantitative reverse transcriptase polymerase chain reaction. The amount of mrjp1 transcript from the isolated bees was normalized to that of the colony bees. Data are expressed as means ± S.D. (n = 3). |
3.3. Expression of mrjp genesin the mushroom bodyand hypopharyngeal gland
The honeybee has nine mrjp genes (Drapeau et al. , 2006), and MRJP proteins are components of the royal jelly synthesized in the hypopharyngeal gland of nurse bees (Ohashi et al. , 1997; Simüth, 2001). Because the hypopharyngeal gland is also located in the head of worker bees and is well-developed in nine-day-old colony bees, mrjp1 expression detected in the mushroom body might be due to contamination of the transcript from the hypopharyngeal gland. To test this possibility, we compared the composition of mrjp transcripts between the mushroom body and hypopharyngeal gland collected from three individual nurse bees by qRT-PCR using primer sets that specifically amplify each of the nine genes (Tab. I). The mrjp7, mrjp1 and mrjp2 transcripts were the three most abundant transcripts in the mushroom body, whereas mrjp1, mrjp3 and mrjp2 were the most abundant transcripts in the hypopharyngeal gland (Fig. 3). The expression ratio of mrjp1 to mrjp3 was 14.7 ± 2.9 (mean ± s.d.) in the mushroom body and 4.4 ± 0.3 in the hypopharyngeal gland, respectively. The difference was significant (one-factor ANOVA, F = 35.7, P = 0.039), suggesting that at least part of mrjp1 transcripts detected in the mushroom body are expressed there, even though the mrjp3 transcripts detected in the mushroom body are contaminants from the hypopharyngeal gland. Similar comparisons also suggested that mrjp2 and mrjp7 are expressed in the mushroom body, the expression of mrjp7 being obvious.
 |
Figure 3 Comparison of the major royal jelly protein (mrjp) gene expression patterns between the mushroom body (A) and the hypopharyngeal gland (B) from the same individual measured by real-time quantitative reverse transcriptase polymerase chain reaction. Standard plasmids were constructed by inserting a fragment of each cDNA into the pT7Blue-2 T-Vector to determine the copy number of mrjp transcripts and the actin transcript. The amount of each mrjp transcript relative to that of actin was calculated, and the relative expression level was further normalized to that of mrjp1. Data are expressed as means ± S.D. (n = 3). |
4. DISCUSSION
We investigated differentially expressed genes in the mushroom bodies of colony and isolated worker bees by differential display. This method is based on PCR amplification of gene fragments and is a highly sensitive method for detecting small differences in gene expression (Liang and Pardee, 1992). Although the results of differential display may lack reproducibility (Livesey and Hunt, 1996), we identified 16 bands that were differentially and reproducibly expressed between the colony and isolated bees using two independent sets of RNA samples. All other bands showed nearly equal intensity between the colony and isolated bees, suggesting that the difference in rearing condition affected the expression of only specific genes. Ichikawa and Sasaki (2003) reported that isolation of worker bees decreased the development of their learning ability. Our data show that isolation also affects the expression of some genes in the mushroom body, suggesting that such changes in gene expression might lead to degradation of brain function. However, the cause of altered gene expression is unclear. We believe that lack of social environment in isolation could be a causal factor. However, it is also possible that poor nutrition affected gene expression in the isolated bees, as they were provided with only sucrose solution.
The mrjp1 gene was the most prominent colony bee-specific band found in the differential display analysis (Fig. 1), and the differential expression of mrjp1 was further confirmed by qRT-PCR (Fig. 2). MRJP1 is a royal jelly protein synthesized in the hypopharyngeal gland and is a component of food for the larvae and queen (Hanes and Simüth, 1992; Klaudiny et al. , 1994; Kubo et al. , 1996; Ohashi et al. , 1997; Simüth, 2001; Santos et al. , 2005). Kucharski et al. (1998) demonstrated that mrjp1 was expressed in the mushroom body and its expression changed depending on the age of the honeybee, suggesting that the expression was correlated with the development of learning and memory. We demonstrated that the mrjp1 expression level in the mushroom body was reduced in isolated bees, further supporting the hypothesis that mrjp1 gene expression is associated with the development of learning ability.
MRJPs share a common evolutionary origin with the Yellow protein family (Albert et al. , 1996, 1999; Maleszka and Kucharski, 2000; Albert and Klaudiny, 2004). Recent genomic analysis of A. mellifera suggests that mrjps evolved from yellow-e3 (Drapeau et al. , 2006). Yellow was originally identified as a gene involved in the pigmentation of larval and adult cuticle in Drosophila melanogaster (Nash, 1976). There are 14 yellow-related proteins in D. melanogaster, and they have diverse functions, including pigmentation, reproductive maturation and regulation of sex-specific behaviours (Drapeau, 2003). The putative function of Drosophila yellow-e3 is in early brain development (Drapeau et al. , 2006). Therefore, it is possible that MRJPs also have regulatory functions that control development and behaviour of honeybees (Maleszka and Kucharski, 2000) in addition to their nutritional functions (Albert et al. , 1999).
Studies have suggested that MRJP1 is a multifunctional protein. Besides its importance as a nutrient in royal jelly, MRJP1 may protect the royal jelly from bacterial infections through the formation of a short digestion product of the MRJP1 C terminus, which has antimicrobial activity (Fontana et al. , 2004). MRJP1 is also present in honey and honeybee pollen, namely in pollen pellets and pollen bread. Although the functions of MRJP1 in these honeybee products are not completely elucidated, it is postulated that the MRJP1 participates in physical–mechanical processing of honey and honeybee pollen (Simüth et al. , 2004, in which MRJP1 is referred to as apalbumin-1). Furthermore, it has been reported that MRJP1 is physiologically active in mammalian cells: The N-terminal fragment of MRJP1 stimulates mouse macrophages to release tumour necrosis factor alpha (Simüth et al. , 2004; Majtan et al. , 2006), and MRJP1 has growth factor-like properties in primary-cultured rat hepatocytes (Kamakura and Sakaki, 2006). Our results, together with those reported by Kucharski et al. (1998), suggests that MRJP1 has a function in the brain and is possibly involved in the development of learning ability. MRJP1 appears to be an important multifunctional protein that is related to the honeybee’s unique character as a social insect.
Acknowledgments
We thank K. Harano, S. Nomura and M. Miyagawa for their help with honeybee sampling. This work was supported by JSPS Research Fellowships from the Japan Society for Young Scientists to M. H., and by a Research Frontier Program of Tamagawa University (F030041) research grant from the Ministry of Education, Culture, Sports, Science and Technology.
References
- Albert S., Klaudiny J. (2004) The MRJP/YELLOW protein family of Apis mellifera: identification of new members in the EST library, J. Insect Physiol. 50, 51–59. [CrossRef] [PubMed] [Google Scholar]
- Albert S., Bhattacharya D., Klaudiny J., Schmitzova J., Simüth J. (1999) The family of major royal jelly proteins and its evolution, J. Mol. Evol. 49, 290–297. [CrossRef] [PubMed] [Google Scholar]
- Albert S., Klaudiny J., Simüth J. (1996) Newly discovered features of the updated sequence of royal jelly protein RJP57-1; longer repetitive region on C-terminus and homology to Drosophila melanogaster yellow protein, J. Apicult. Res. 35, 63–68. [Google Scholar]
- Applied Biosystems (2001) Relative quantification of gene expression, 7700 Sequence Detection System User Bulletin 2. [Google Scholar]
- Barchuk A.R., Cristino A.S., Kucharski R., Costa L.F., Simoes Z.L., Maleszka R. (2007) Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera, BMC Dev. Biol. 7, 70. [Google Scholar]
- Bitterman M.E., Menzel R., Fietz A., Schafer S. (1983) Classical conditioning of proboscis extension in honeybees (Apis mellifera), J. Comp. Psychol. 97, 107–119. [CrossRef] [PubMed] [Google Scholar]
- Cash A.C., Whitfield C.W., Ismail N., Robinson G.E. (2005) Behavior and the limits of genomic plasticity: power and replicability in microarray analysis of honeybee brains, Genes Brain Behav. 4, 267–271. [CrossRef] [PubMed] [Google Scholar]
- Chen Y.P., Higgins J.A., Feldlaufer M.F. (2005) Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.), Appl. Environ. Microb. 71, 436–441. [Google Scholar]
- Drapeau M.D. (2003) A novel hypothesis on the biochemical role of the Drosophila Yellow protein, Biochem. Bioph. Res. Co. 311, 1–3. [Google Scholar]
- Drapeau M.D., Albert S., Kucharski R., Prusko C., Maleszka R. (2006) Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees, Genome Res. 16, 1385–1394. [CrossRef] [PubMed] [Google Scholar]
- Durst C., Eichmuller S., Menzel R. (1994) Development and experience lead to increased volume of subcompartments of the honeybee mushroom body, Behav. Neural. Biol. 62, 259–263. [CrossRef] [PubMed] [Google Scholar]
- Erber J., Masuhr T.H., Menzel R. (1980) Localization of short-term memory in the brain of the bee, Apis mellifera, Physiol. Entomol. 5, 343–358. [CrossRef] [Google Scholar]
- Evans J.D., Wheeler D.E. (2001) Gene expression and the evolution of insect polyphenisms, Bioessays 23, 62–68. [CrossRef] [PubMed] [Google Scholar]
- Fahrbach S.E., Robinson G.E. (1995) Behavioral development in the honey bee: toward the study of learning under natural conditions, Learn. Memory 2, 199–224. [CrossRef] [Google Scholar]
- Fontana R., Mendes M.A., de Souza B.M., Konno K., Cesar L.M., Malaspina O., Palma M.S. (2004) Jelleines: a family of antimicrobial peptides from the Royal Jelly of honeybees (Apis mellifera), Peptides 25, 919–928. [CrossRef] [PubMed] [Google Scholar]
- Frisch K. von (1967) Honeybees: do they use direction and distance information provided by their dancers? Science 158, 1072–1076. [CrossRef] [PubMed] [Google Scholar]
- Hammer M., Menzel R. (1995) Learning and memory in the honeybee, J. Neurosci. 15, 1617–1630. [PubMed] [Google Scholar]
- Hanes J., Simüth J. (1992) Identification and partial characterization of the major royal jelly protein of the honey bee (Apis mellifera L.), J. Apicult. Res. 31, 22–26. [Google Scholar]
- Heisenberg M. (1998) What do the mushroom bodies do for the insect brain? Learn. Memory 5, 1–10. [Google Scholar]
- The Honey Bee Genome Sequencing Consortium (2006) Insights into social insects from the genome of the honey bee Apis mellifera, Nature 443, 931–949. [Google Scholar]
- Ichikawa N., Sasaki M. (2003) Importance of social stimuli for the development of learning capability in honeybees, Appl. Entomol. Zool. 38, 203–209. [CrossRef] [Google Scholar]
- Kamakura M., Sakaki T. (2006) A hypopharyngeal gland protein of the worker honeybee Apis mellifera L. enhances proliferation of primary-cultured rat hepatocytes and suppresses apoptosis in the absence of serum, Protein Expres. Purif. 45, 307–314. [CrossRef] [Google Scholar]
- Kamikouchi A., Takeuchi H., Sawata M., Natori S., Kubo T. (2000) Concentrated expression of Ca2+/calmodulin-dependent protein kinase II and protein kinase C in the mushroom bodies of the brain of the honeybee Apis mellifera L., J. Comp. Neurol. 417, 501–510. [CrossRef] [PubMed] [Google Scholar]
- Kamikouchi A., Takeuchi H., Sawata M., Ohashi K., Natori S., Kubo T. (1998) Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homologue in the mushroom bodies of the brain of the worker honeybee Apis mellifera L., Biochem. Biophys. Res. Co. 242, 181–186. [CrossRef] [Google Scholar]
- Klaudiny J., Kulifajova J., Crailsheim K., Simuth J. (1994) New approach to the study of division of labour in the honeybee colony (Apis mellifera L.), Apidologie 25, 596–600. [CrossRef] [EDP Sciences] [Google Scholar]
- Kubo T., Sasaki M., Nakamura J., Sasagawa H., Ohashi K., Takeuchi H., Natori S. (1996) Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role, J. Biochem. 119, 291–295. [PubMed] [Google Scholar]
- Kucharski R., Maleszka R., Hayward D.C., Ball E.E. (1998) A royal jelly protein is expressed in a subset of Kenyon cells in the mushroom bodies of the honey bee brain, Naturwissenschaften 85, 343–346. [CrossRef] [PubMed] [Google Scholar]
- Liang P., Pardee A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction, Science 257, 967–971. [CrossRef] [PubMed] [Google Scholar]
- Livesey F.J., Hunt S.P. (1996) Identifying changes in gene expression in the nervous system: mRNA differential display, Trends Neurosci. 19, 84–88. [CrossRef] [PubMed] [Google Scholar]
- Majtan J., Kovacova E., Bilikova K., Simuth J. (2006) The immunostimulatory effect of the recombinant apalbumin 1-major honeybee royal jelly protein-on TNFalpha release, Int. Immunopharmacol. 6, 269–278. [CrossRef] [PubMed] [Google Scholar]
- Maleszka R., Kucharski R. (2000) Analysis of Drosophila yellow-B cDNA reveals a new family of proteins related to the Royal Jelly proteins in the honeybee and to an orphan protein in an unusual bacterium Deinococcus radiodurans, Biochem. Biophys. Res. Co. 270, 773–776. [CrossRef] [Google Scholar]
- Meller V.H., Davis R.L. (1996) Biochemistry of insect learning: lessons from bees and flies, Insect Biochem. Molec. 26, 327–335. [CrossRef] [Google Scholar]
- Menzel R. (2001) Searching for the memory trace in a mini-brain, the honeybee, Learn. Memory 8, 53–62. [CrossRef] [Google Scholar]
- Menzel R., Giurfa M. (2001) Cognitive architecture of a mini-brain: the honeybee, Trends Cogn. Sci. 5, 62–71. [CrossRef] [PubMed] [Google Scholar]
- Menzel R., Müller U. (1996) Learning and memory in honeybees: from behavior to neural substrates, Annu. Rev. Neurosci. 19, 379–404. [CrossRef] [PubMed] [Google Scholar]
- Nash W.G. (1976) Patterns of pigmentation color states regulated by the y locus in Drosophila melanogaster, Dev. Biol. 48, 336–343. [CrossRef] [PubMed] [Google Scholar]
- Ohashi K., Natori S., Kubo T. (1997) Change in the mode of gene expression of the hypopharyngeal gland cells with an age-dependent role change of the worker honeybee Apis mellifera L., Eur. J. Biochem. 249, 797–802. [CrossRef] [PubMed] [Google Scholar]
- Robinson G.E. (1992) Regulation of division of labor in insect societies, Annu. Rev. Entomol. 37, 637–665. [CrossRef] [PubMed] [Google Scholar]
- Robinson G.E., Fahrbach S.E., Winston M.L. (1997) Insect societies and the molecular biology of social behavior, Bioessays 19, 1099–1108. [CrossRef] [PubMed] [Google Scholar]
- Robinson G.E., Grozinger C.M., Whitfield C.W. (2005) Sociogenomics: social life in molecular terms, Nat. Rev. Genet. 6, 257–270. [CrossRef] [PubMed] [Google Scholar]
- Rybak J., Menzel R. (1998) Integrative properties of the Pe1 neuron, a unique mushroom body output neuron, Learn. Memory 5, 133–145. [Google Scholar]
- Santos K.S., dos Santos L.D., Mendes M.A., de Souza B.M., Malaspina O., Palma M.S. (2005) Profiling the proteome complement of the secretion from hypopharyngeal gland of Africanized nurse-honeybees (Apis mellifera L.), Insect Biochem. Molec. 35, 85–91. [CrossRef] [Google Scholar]
- Sawata M., Yoshino D., Takeuchi H., Kamikouchi A., Ohashi K., Kubo T. (2002) Identi?cation and punctate nuclear localization of a novel noncoding RNA, Ks-1, from the honeybee brain, RNA 8, 772–785. [CrossRef] [PubMed] [Google Scholar]
- Simüth J. (2001) Some properties of the main protein of honeybee (Apis mellifera) royal jelly, Apidologie 32, 69–80. [CrossRef] [EDP Sciences] [Google Scholar]
- Simüth J., Bilikova K., Kovacova E., Kuzmova Z., Schroder W. (2004) Immunochemical approach to detection of adulteration in honey: physiologically active royal jelly protein stimulating TNF-alpha release is a regular component of honey, J. Agr. Food Chem. 52, 2154–2158. [CrossRef] [Google Scholar]
- Takeuchi H., Fujiyuki T., Shirai K., Matsuo Y., Kamikouchi A., Fujinawa Y., Kato A., Tsujimoto A., Kubo T. (2002) Identification of genes expressed referentially in the honeybee mushroom bodies by combination of differential display and cDNA microarray, FEBS Lett. 513, 230–234. [CrossRef] [PubMed] [Google Scholar]
- Takeuchi H., Kage E., Sawata M., Kamikouchi A., Ohashi K., Ohara M., Fujiyuki T., Kunieda T., Sekimizu K., Natori S., Kubo T. (2001) Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honeybee brain, Insect Mol. Biol. 10, 487–494. [CrossRef] [PubMed] [Google Scholar]
- Takeuchi H., Yasuda A., Yasuda-Kamatani Y., Sawata M., Matsuo Y., Kato A., Tsujimoto A., Nakajima T., Kubo T. (2004) Prepro-tachykinin gene expression in the brain of the honeybee Apis mellifera, Cell Tissue Res. 316, 281–293. [CrossRef] [PubMed] [Google Scholar]
- Thompson G.J., Kucharski R., Maleszka R., Oldroyd B.P. (2006) Towards a molecular definition of worker sterility: differential gene expression and reproductive plasticity in honey bees, Insect Mol. Biol. 15, 637–644. [PubMed] [Google Scholar]
- Whitfield C.W., Band M.R., Bonaldo M.F., Kumar C.G., Liu L., Pardinas J.R., Robertson H.M., Soares M.B., Robinson G.E. (2002) Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee, Genome Res. 12, 555–566. [CrossRef] [PubMed] [Google Scholar]
- Whitfield C.W., Cziko A.M., Robinson G.E. (2003) Gene expression profiles in the brain predict behavior in individual honey bees, Science 302, 296–299. [CrossRef] [PubMed] [Google Scholar]
- Whitfield C.W., Ben-Shahar Y., Brillet C., Leoncini I., Crauser D., Leconte Y., Rodriguez-Zas S., Robinson G.E. (2006) Genomic dissection of behavioral maturation in the honey bee, Proc. Natl Acad. Sci. (USA) 103, 16068–16075. [Google Scholar]
- Winston M.L. (1987) The Biology of the Honeybee, Harvard Univ. Press, Cambridge, pp. 1–281. [Google Scholar]
- Yamazaki Y., Shirai K., Paul R.K., Fujiyuki T., Wakamoto A., Takeuchi H., Kubo T. (2006) Differential expression of HR38 in the mushroom bodies of the honeybee brain depends on the caste and division of labor, FEBS Lett. 580, 2667–2670. [CrossRef] [PubMed] [Google Scholar]
All Tables
Primer sets of major royal jelly protein (mrjp) genes for real-time quantitative reverse transcriptase polymerase chain reaction.
All Figures
 |
Figure 1 An electrophoresis profile of the fluorescent differential display comparing gene expression in the mushroom bodies of colony bees (C+) and isolated bees (I+). Numbers indicate molecular size (bp). Arrowhead indicates approximately 1.1-kbp band of the colony bee-specific gene. No bands were detected in the negative (RT-) control lanes (C- and I-). |
| In the text | |
 |
Figure 2 Relative major royal jelly protein (mrjp) 1 gene expression level measured by real-time quantitative reverse transcriptase polymerase chain reaction. The amount of mrjp1 transcript from the isolated bees was normalized to that of the colony bees. Data are expressed as means ± S.D. (n = 3). |
| In the text | |
 |
Figure 3 Comparison of the major royal jelly protein (mrjp) gene expression patterns between the mushroom body (A) and the hypopharyngeal gland (B) from the same individual measured by real-time quantitative reverse transcriptase polymerase chain reaction. Standard plasmids were constructed by inserting a fragment of each cDNA into the pT7Blue-2 T-Vector to determine the copy number of mrjp transcripts and the actin transcript. The amount of each mrjp transcript relative to that of actin was calculated, and the relative expression level was further normalized to that of mrjp1. Data are expressed as means ± S.D. (n = 3). |
| In the text | |


